Collapse reshape benchmark
data.table is an R package
for efficient data manipulation. I have an NSF POSE grant about
expanding the open-source ecosystem of users and contributors around
data.table. Part of that project is benchmarking time and memory
usage, and comparing with similar packages. Similar to previous posts
about
reshaping
and
reading/writing/summarization,
the goal of this post is to explain how to use
atime to benchmark data.table
reshape with similar packages.
Background about data reshaping
Data reshaping means changing the shape of the data, in order to get it into a more appropriate format, for learning/plotting/etc. It is perhaps best explained using a simple example. Here we consider the iris data, which has four numeric columns, and we show the first two rows below:
(two.iris.wide <- iris[c(1,150),])
## Sepal.Length Sepal.Width Petal.Length Petal.Width Species
## 1 5.1 3.5 1.4 0.2 setosa
## 150 5.9 3.0 5.1 1.8 virginica
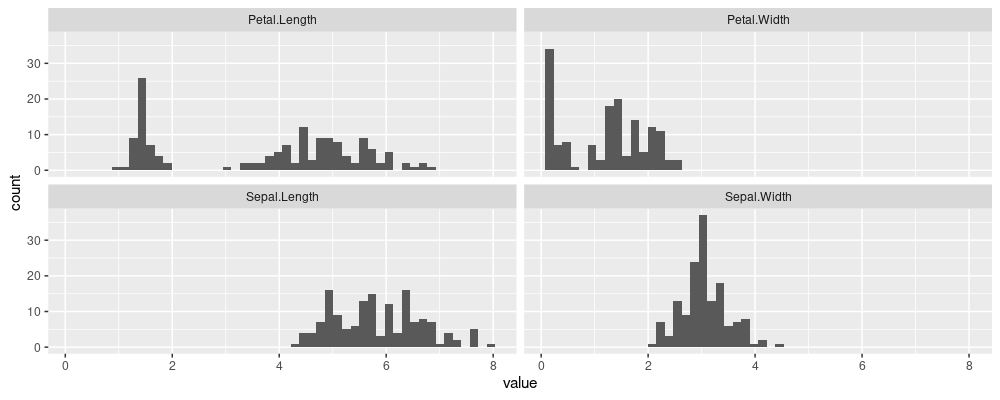
Note the table above has 8 numbers, arranged into a table of 2 rows and 4 columns. What if we wanted to make a facetted histogram of the numeric iris data columns, with one panel/facet for each column? With ggplots we have to first reshape to “long” (or I like to say “tall”) format:
cols.to.reshape <- c("Sepal.Length","Sepal.Width","Petal.Length","Petal.Width")
two.iris.wide.mat <- as.matrix(two.iris.wide[,cols.to.reshape])
row.i <- as.integer(row(two.iris.wide.mat))
(two.iris.tall <- data.frame(
row.i,
Species=two.iris.wide$Species[row.i],
row.name=rownames(two.iris.wide)[row.i],
col.name=cols.to.reshape[as.integer(col(two.iris.wide.mat))],
cm=as.numeric(two.iris.wide.mat)))
## row.i Species row.name col.name cm
## 1 1 setosa 1 Sepal.Length 5.1
## 2 2 virginica 150 Sepal.Length 5.9
## 3 1 setosa 1 Sepal.Width 3.5
## 4 2 virginica 150 Sepal.Width 3.0
## 5 1 setosa 1 Petal.Length 1.4
## 6 2 virginica 150 Petal.Length 5.1
## 7 1 setosa 1 Petal.Width 0.2
## 8 2 virginica 150 Petal.Width 1.8
Note the table above has the same 8 numbers, but arranged into a table of 8 rows and 1 column, which is the desired input for ggplots.
Making a function
Here is how we would do it using a function:
reshape_taller <- function(DF, col.i){
orig.row.i <- 1:nrow(DF)
wide.mat <- as.matrix(DF[, col.i])
orig.col.i <- as.integer(col(wide.mat))
other.values <- DF[, -col.i, drop=FALSE]
rownames(other.values) <- NULL
data.frame(
orig.row.i,
orig.row.name=rownames(DF)[orig.row.i],
orig.col.i,
orig.col.name=names(DF)[orig.col.i],
other.values,
value=as.numeric(wide.mat))
}
reshape_taller(two.iris.wide,1:4)
## orig.row.i orig.row.name orig.col.i orig.col.name Species value
## 1 1 1 1 Sepal.Length setosa 5.1
## 2 2 150 1 Sepal.Length virginica 5.9
## 3 1 1 2 Sepal.Width setosa 3.5
## 4 2 150 2 Sepal.Width virginica 3.0
## 5 1 1 3 Petal.Length setosa 1.4
## 6 2 150 3 Petal.Length virginica 5.1
## 7 1 1 4 Petal.Width setosa 0.2
## 8 2 150 4 Petal.Width virginica 1.8
And below is with the full iris data set:
iris.tall <- reshape_taller(iris,1:4)
library(ggplot2)
ggplot()+
geom_histogram(aes(
value),
bins=50,
data=iris.tall)+
facet_wrap("orig.col.name")
Comparison with other functions
The function defined above works, and there are other functions which provide similar functionality. For example,
head(stats::reshape(
iris, direction="long", varying=list(cols.to.reshape), v.names="cm"))
## Species time cm id
## 1.1 setosa 1 5.1 1
## 2.1 setosa 1 4.9 2
## 3.1 setosa 1 4.7 3
## 4.1 setosa 1 4.6 4
## 5.1 setosa 1 5.0 5
## 6.1 setosa 1 5.4 6
library(data.table)
## data.table 1.16.99 EN DEVELOPPEMENT build 2024-09-23 11:12:40 UTC utilisant 1 threads (voir ?getDTthreads). Dernières actualités : r-datatable.com
## **********
## Exécution de data.table en anglais ; L'aide du package n'est disponible qu'en anglais. Lorsque vous recherchez de l'aide en ligne, veillez à vérifier également le message d'erreur en anglais. Pour ce faire, consultez les fichiers po/R-<locale>.po et po/<locale>.po dans le source du package, où les messages d'erreur en langue native et en anglais sont mis côte à côte
## **********
iris.dt <- data.table(iris)
melt(iris.dt, measure.vars=cols.to.reshape, value.name="cm")
## Species variable cm
## <fctr> <fctr> <num>
## 1: setosa Sepal.Length 5.1
## 2: setosa Sepal.Length 4.9
## 3: setosa Sepal.Length 4.7
## 4: setosa Sepal.Length 4.6
## 5: setosa Sepal.Length 5.0
## ---
## 596: virginica Petal.Width 2.3
## 597: virginica Petal.Width 1.9
## 598: virginica Petal.Width 2.0
## 599: virginica Petal.Width 2.3
## 600: virginica Petal.Width 1.8
tidyr::pivot_longer(iris, cols.to.reshape, values_to = "cm")
## Warning: Using an external vector in selections was deprecated in tidyselect 1.1.0.
## ℹ Please use `all_of()` or `any_of()` instead.
## # Was:
## data %>% select(cols.to.reshape)
##
## # Now:
## data %>% select(all_of(cols.to.reshape))
##
## See <https://tidyselect.r-lib.org/reference/faq-external-vector.html>.
## This warning is displayed once every 8 hours.
## Call `lifecycle::last_lifecycle_warnings()` to see where this warning was
## generated.
## # A tibble: 600 × 3
## Species name cm
## <fct> <chr> <dbl>
## 1 setosa Sepal.Length 5.1
## 2 setosa Sepal.Width 3.5
## 3 setosa Petal.Length 1.4
## 4 setosa Petal.Width 0.2
## 5 setosa Sepal.Length 4.9
## 6 setosa Sepal.Width 3
## 7 setosa Petal.Length 1.4
## 8 setosa Petal.Width 0.2
## 9 setosa Sepal.Length 4.7
## 10 setosa Sepal.Width 3.2
## # ℹ 590 more rows
head(collapse::pivot(iris, values=cols.to.reshape, names=list("variable", "cm")))
## Species variable cm
## 1 setosa Sepal.Length 5.1
## 2 setosa Sepal.Length 4.9
## 3 setosa Sepal.Length 4.7
## 4 setosa Sepal.Length 4.6
## 5 setosa Sepal.Length 5.0
## 6 setosa Sepal.Length 5.4
polars::as_polars_df(iris)$unpivot(index="Species", value_name="cm")
<div><style>
.dataframe > thead > tr > th,
.dataframe > tbody > tr > td {
text-align: right;
white-space: pre-wrap;
}
</style>
<small>shape: (600, 3)</small><table border="1" class="dataframe"><thead><tr><th>Species</th><th>variable</th><th>cm</th></tr><tr><td>cat</td><td>str</td><td>f64</td></tr></thead><tbody><tr><td>"setosa"</td><td>"Sepal.Length"</td><td>5.1</td></tr><tr><td>"setosa"</td><td>"Sepal.Length"</td><td>4.9</td></tr><tr><td>"setosa"</td><td>"Sepal.Length"</td><td>4.7</td></tr><tr><td>"setosa"</td><td>"Sepal.Length"</td><td>4.6</td></tr><tr><td>"setosa"</td><td>"Sepal.Length"</td><td>5.0</td></tr><tr><td>…</td><td>…</td><td>…</td></tr><tr><td>"virginica"</td><td>"Petal.Width"</td><td>2.3</td></tr><tr><td>"virginica"</td><td>"Petal.Width"</td><td>1.9</td></tr><tr><td>"virginica"</td><td>"Petal.Width"</td><td>2.0</td></tr><tr><td>"virginica"</td><td>"Petal.Width"</td><td>2.3</td></tr><tr><td>"virginica"</td><td>"Petal.Width"</td><td>1.8</td></tr></tbody></table></div>
con <- duckdb::dbConnect(duckdb::duckdb(), dbdir = ":memory:")
duckdb::dbWriteTable(con, "iris_table", iris)
DBI::dbGetQuery(con, 'UNPIVOT iris_table ON "Sepal.Length", "Petal.Length", "Sepal.Width", "Petal.Width" INTO NAME part_dim VALUE cm')
## Species part_dim cm
## 1 setosa Sepal.Length 5.1
## 2 setosa Petal.Length 1.4
## 3 setosa Sepal.Width 3.5
## 4 setosa Petal.Width 0.2
## 5 setosa Sepal.Length 4.9
## 6 setosa Petal.Length 1.4
## 7 setosa Sepal.Width 3.0
## 8 setosa Petal.Width 0.2
## 9 setosa Sepal.Length 4.7
## 10 setosa Petal.Length 1.3
## 11 setosa Sepal.Width 3.2
## 12 setosa Petal.Width 0.2
## 13 setosa Sepal.Length 4.6
## 14 setosa Petal.Length 1.5
## 15 setosa Sepal.Width 3.1
## 16 setosa Petal.Width 0.2
## 17 setosa Sepal.Length 5.0
## 18 setosa Petal.Length 1.4
## 19 setosa Sepal.Width 3.6
## 20 setosa Petal.Width 0.2
## 21 setosa Sepal.Length 5.4
## 22 setosa Petal.Length 1.7
## 23 setosa Sepal.Width 3.9
## 24 setosa Petal.Width 0.4
## 25 setosa Sepal.Length 4.6
## 26 setosa Petal.Length 1.4
## 27 setosa Sepal.Width 3.4
## 28 setosa Petal.Width 0.3
## 29 setosa Sepal.Length 5.0
## 30 setosa Petal.Length 1.5
## 31 setosa Sepal.Width 3.4
## 32 setosa Petal.Width 0.2
## 33 setosa Sepal.Length 4.4
## 34 setosa Petal.Length 1.4
## 35 setosa Sepal.Width 2.9
## 36 setosa Petal.Width 0.2
## 37 setosa Sepal.Length 4.9
## 38 setosa Petal.Length 1.5
## 39 setosa Sepal.Width 3.1
## 40 setosa Petal.Width 0.1
## 41 setosa Sepal.Length 5.4
## 42 setosa Petal.Length 1.5
## 43 setosa Sepal.Width 3.7
## 44 setosa Petal.Width 0.2
## 45 setosa Sepal.Length 4.8
## 46 setosa Petal.Length 1.6
## 47 setosa Sepal.Width 3.4
## 48 setosa Petal.Width 0.2
## 49 setosa Sepal.Length 4.8
## 50 setosa Petal.Length 1.4
## 51 setosa Sepal.Width 3.0
## 52 setosa Petal.Width 0.1
## 53 setosa Sepal.Length 4.3
## 54 setosa Petal.Length 1.1
## 55 setosa Sepal.Width 3.0
## 56 setosa Petal.Width 0.1
## 57 setosa Sepal.Length 5.8
## 58 setosa Petal.Length 1.2
## 59 setosa Sepal.Width 4.0
## 60 setosa Petal.Width 0.2
## 61 setosa Sepal.Length 5.7
## 62 setosa Petal.Length 1.5
## 63 setosa Sepal.Width 4.4
## 64 setosa Petal.Width 0.4
## 65 setosa Sepal.Length 5.4
## 66 setosa Petal.Length 1.3
## 67 setosa Sepal.Width 3.9
## 68 setosa Petal.Width 0.4
## 69 setosa Sepal.Length 5.1
## 70 setosa Petal.Length 1.4
## 71 setosa Sepal.Width 3.5
## 72 setosa Petal.Width 0.3
## 73 setosa Sepal.Length 5.7
## 74 setosa Petal.Length 1.7
## 75 setosa Sepal.Width 3.8
## 76 setosa Petal.Width 0.3
## 77 setosa Sepal.Length 5.1
## 78 setosa Petal.Length 1.5
## 79 setosa Sepal.Width 3.8
## 80 setosa Petal.Width 0.3
## 81 setosa Sepal.Length 5.4
## 82 setosa Petal.Length 1.7
## 83 setosa Sepal.Width 3.4
## 84 setosa Petal.Width 0.2
## 85 setosa Sepal.Length 5.1
## 86 setosa Petal.Length 1.5
## 87 setosa Sepal.Width 3.7
## 88 setosa Petal.Width 0.4
## 89 setosa Sepal.Length 4.6
## 90 setosa Petal.Length 1.0
## 91 setosa Sepal.Width 3.6
## 92 setosa Petal.Width 0.2
## 93 setosa Sepal.Length 5.1
## 94 setosa Petal.Length 1.7
## 95 setosa Sepal.Width 3.3
## 96 setosa Petal.Width 0.5
## 97 setosa Sepal.Length 4.8
## 98 setosa Petal.Length 1.9
## 99 setosa Sepal.Width 3.4
## 100 setosa Petal.Width 0.2
## 101 setosa Sepal.Length 5.0
## 102 setosa Petal.Length 1.6
## 103 setosa Sepal.Width 3.0
## 104 setosa Petal.Width 0.2
## 105 setosa Sepal.Length 5.0
## 106 setosa Petal.Length 1.6
## 107 setosa Sepal.Width 3.4
## 108 setosa Petal.Width 0.4
## 109 setosa Sepal.Length 5.2
## 110 setosa Petal.Length 1.5
## 111 setosa Sepal.Width 3.5
## 112 setosa Petal.Width 0.2
## 113 setosa Sepal.Length 5.2
## 114 setosa Petal.Length 1.4
## 115 setosa Sepal.Width 3.4
## 116 setosa Petal.Width 0.2
## 117 setosa Sepal.Length 4.7
## 118 setosa Petal.Length 1.6
## 119 setosa Sepal.Width 3.2
## 120 setosa Petal.Width 0.2
## 121 setosa Sepal.Length 4.8
## 122 setosa Petal.Length 1.6
## 123 setosa Sepal.Width 3.1
## 124 setosa Petal.Width 0.2
## 125 setosa Sepal.Length 5.4
## 126 setosa Petal.Length 1.5
## 127 setosa Sepal.Width 3.4
## 128 setosa Petal.Width 0.4
## 129 setosa Sepal.Length 5.2
## 130 setosa Petal.Length 1.5
## 131 setosa Sepal.Width 4.1
## 132 setosa Petal.Width 0.1
## 133 setosa Sepal.Length 5.5
## 134 setosa Petal.Length 1.4
## 135 setosa Sepal.Width 4.2
## 136 setosa Petal.Width 0.2
## 137 setosa Sepal.Length 4.9
## 138 setosa Petal.Length 1.5
## 139 setosa Sepal.Width 3.1
## 140 setosa Petal.Width 0.2
## 141 setosa Sepal.Length 5.0
## 142 setosa Petal.Length 1.2
## 143 setosa Sepal.Width 3.2
## 144 setosa Petal.Width 0.2
## 145 setosa Sepal.Length 5.5
## 146 setosa Petal.Length 1.3
## 147 setosa Sepal.Width 3.5
## 148 setosa Petal.Width 0.2
## 149 setosa Sepal.Length 4.9
## 150 setosa Petal.Length 1.4
## 151 setosa Sepal.Width 3.6
## 152 setosa Petal.Width 0.1
## 153 setosa Sepal.Length 4.4
## 154 setosa Petal.Length 1.3
## 155 setosa Sepal.Width 3.0
## 156 setosa Petal.Width 0.2
## 157 setosa Sepal.Length 5.1
## 158 setosa Petal.Length 1.5
## 159 setosa Sepal.Width 3.4
## 160 setosa Petal.Width 0.2
## 161 setosa Sepal.Length 5.0
## 162 setosa Petal.Length 1.3
## 163 setosa Sepal.Width 3.5
## 164 setosa Petal.Width 0.3
## 165 setosa Sepal.Length 4.5
## 166 setosa Petal.Length 1.3
## 167 setosa Sepal.Width 2.3
## 168 setosa Petal.Width 0.3
## 169 setosa Sepal.Length 4.4
## 170 setosa Petal.Length 1.3
## 171 setosa Sepal.Width 3.2
## 172 setosa Petal.Width 0.2
## 173 setosa Sepal.Length 5.0
## 174 setosa Petal.Length 1.6
## 175 setosa Sepal.Width 3.5
## 176 setosa Petal.Width 0.6
## 177 setosa Sepal.Length 5.1
## 178 setosa Petal.Length 1.9
## 179 setosa Sepal.Width 3.8
## 180 setosa Petal.Width 0.4
## 181 setosa Sepal.Length 4.8
## 182 setosa Petal.Length 1.4
## 183 setosa Sepal.Width 3.0
## 184 setosa Petal.Width 0.3
## 185 setosa Sepal.Length 5.1
## 186 setosa Petal.Length 1.6
## 187 setosa Sepal.Width 3.8
## 188 setosa Petal.Width 0.2
## 189 setosa Sepal.Length 4.6
## 190 setosa Petal.Length 1.4
## 191 setosa Sepal.Width 3.2
## 192 setosa Petal.Width 0.2
## 193 setosa Sepal.Length 5.3
## 194 setosa Petal.Length 1.5
## 195 setosa Sepal.Width 3.7
## 196 setosa Petal.Width 0.2
## 197 setosa Sepal.Length 5.0
## 198 setosa Petal.Length 1.4
## 199 setosa Sepal.Width 3.3
## 200 setosa Petal.Width 0.2
## 201 versicolor Sepal.Length 7.0
## 202 versicolor Petal.Length 4.7
## 203 versicolor Sepal.Width 3.2
## 204 versicolor Petal.Width 1.4
## 205 versicolor Sepal.Length 6.4
## 206 versicolor Petal.Length 4.5
## 207 versicolor Sepal.Width 3.2
## 208 versicolor Petal.Width 1.5
## 209 versicolor Sepal.Length 6.9
## 210 versicolor Petal.Length 4.9
## 211 versicolor Sepal.Width 3.1
## 212 versicolor Petal.Width 1.5
## 213 versicolor Sepal.Length 5.5
## 214 versicolor Petal.Length 4.0
## 215 versicolor Sepal.Width 2.3
## 216 versicolor Petal.Width 1.3
## 217 versicolor Sepal.Length 6.5
## 218 versicolor Petal.Length 4.6
## 219 versicolor Sepal.Width 2.8
## 220 versicolor Petal.Width 1.5
## 221 versicolor Sepal.Length 5.7
## 222 versicolor Petal.Length 4.5
## 223 versicolor Sepal.Width 2.8
## 224 versicolor Petal.Width 1.3
## 225 versicolor Sepal.Length 6.3
## 226 versicolor Petal.Length 4.7
## 227 versicolor Sepal.Width 3.3
## 228 versicolor Petal.Width 1.6
## 229 versicolor Sepal.Length 4.9
## 230 versicolor Petal.Length 3.3
## 231 versicolor Sepal.Width 2.4
## 232 versicolor Petal.Width 1.0
## 233 versicolor Sepal.Length 6.6
## 234 versicolor Petal.Length 4.6
## 235 versicolor Sepal.Width 2.9
## 236 versicolor Petal.Width 1.3
## 237 versicolor Sepal.Length 5.2
## 238 versicolor Petal.Length 3.9
## 239 versicolor Sepal.Width 2.7
## 240 versicolor Petal.Width 1.4
## 241 versicolor Sepal.Length 5.0
## 242 versicolor Petal.Length 3.5
## 243 versicolor Sepal.Width 2.0
## 244 versicolor Petal.Width 1.0
## 245 versicolor Sepal.Length 5.9
## 246 versicolor Petal.Length 4.2
## 247 versicolor Sepal.Width 3.0
## 248 versicolor Petal.Width 1.5
## 249 versicolor Sepal.Length 6.0
## 250 versicolor Petal.Length 4.0
## 251 versicolor Sepal.Width 2.2
## 252 versicolor Petal.Width 1.0
## 253 versicolor Sepal.Length 6.1
## 254 versicolor Petal.Length 4.7
## 255 versicolor Sepal.Width 2.9
## 256 versicolor Petal.Width 1.4
## 257 versicolor Sepal.Length 5.6
## 258 versicolor Petal.Length 3.6
## 259 versicolor Sepal.Width 2.9
## 260 versicolor Petal.Width 1.3
## 261 versicolor Sepal.Length 6.7
## 262 versicolor Petal.Length 4.4
## 263 versicolor Sepal.Width 3.1
## 264 versicolor Petal.Width 1.4
## 265 versicolor Sepal.Length 5.6
## 266 versicolor Petal.Length 4.5
## 267 versicolor Sepal.Width 3.0
## 268 versicolor Petal.Width 1.5
## 269 versicolor Sepal.Length 5.8
## 270 versicolor Petal.Length 4.1
## 271 versicolor Sepal.Width 2.7
## 272 versicolor Petal.Width 1.0
## 273 versicolor Sepal.Length 6.2
## 274 versicolor Petal.Length 4.5
## 275 versicolor Sepal.Width 2.2
## 276 versicolor Petal.Width 1.5
## 277 versicolor Sepal.Length 5.6
## 278 versicolor Petal.Length 3.9
## 279 versicolor Sepal.Width 2.5
## 280 versicolor Petal.Width 1.1
## 281 versicolor Sepal.Length 5.9
## 282 versicolor Petal.Length 4.8
## 283 versicolor Sepal.Width 3.2
## 284 versicolor Petal.Width 1.8
## 285 versicolor Sepal.Length 6.1
## 286 versicolor Petal.Length 4.0
## 287 versicolor Sepal.Width 2.8
## 288 versicolor Petal.Width 1.3
## 289 versicolor Sepal.Length 6.3
## 290 versicolor Petal.Length 4.9
## 291 versicolor Sepal.Width 2.5
## 292 versicolor Petal.Width 1.5
## 293 versicolor Sepal.Length 6.1
## 294 versicolor Petal.Length 4.7
## 295 versicolor Sepal.Width 2.8
## 296 versicolor Petal.Width 1.2
## 297 versicolor Sepal.Length 6.4
## 298 versicolor Petal.Length 4.3
## 299 versicolor Sepal.Width 2.9
## 300 versicolor Petal.Width 1.3
## 301 versicolor Sepal.Length 6.6
## 302 versicolor Petal.Length 4.4
## 303 versicolor Sepal.Width 3.0
## 304 versicolor Petal.Width 1.4
## 305 versicolor Sepal.Length 6.8
## 306 versicolor Petal.Length 4.8
## 307 versicolor Sepal.Width 2.8
## 308 versicolor Petal.Width 1.4
## 309 versicolor Sepal.Length 6.7
## 310 versicolor Petal.Length 5.0
## 311 versicolor Sepal.Width 3.0
## 312 versicolor Petal.Width 1.7
## 313 versicolor Sepal.Length 6.0
## 314 versicolor Petal.Length 4.5
## 315 versicolor Sepal.Width 2.9
## 316 versicolor Petal.Width 1.5
## 317 versicolor Sepal.Length 5.7
## 318 versicolor Petal.Length 3.5
## 319 versicolor Sepal.Width 2.6
## 320 versicolor Petal.Width 1.0
## 321 versicolor Sepal.Length 5.5
## 322 versicolor Petal.Length 3.8
## 323 versicolor Sepal.Width 2.4
## 324 versicolor Petal.Width 1.1
## 325 versicolor Sepal.Length 5.5
## 326 versicolor Petal.Length 3.7
## 327 versicolor Sepal.Width 2.4
## 328 versicolor Petal.Width 1.0
## 329 versicolor Sepal.Length 5.8
## 330 versicolor Petal.Length 3.9
## 331 versicolor Sepal.Width 2.7
## 332 versicolor Petal.Width 1.2
## 333 versicolor Sepal.Length 6.0
## 334 versicolor Petal.Length 5.1
## 335 versicolor Sepal.Width 2.7
## 336 versicolor Petal.Width 1.6
## 337 versicolor Sepal.Length 5.4
## 338 versicolor Petal.Length 4.5
## 339 versicolor Sepal.Width 3.0
## 340 versicolor Petal.Width 1.5
## 341 versicolor Sepal.Length 6.0
## 342 versicolor Petal.Length 4.5
## 343 versicolor Sepal.Width 3.4
## 344 versicolor Petal.Width 1.6
## 345 versicolor Sepal.Length 6.7
## 346 versicolor Petal.Length 4.7
## 347 versicolor Sepal.Width 3.1
## 348 versicolor Petal.Width 1.5
## 349 versicolor Sepal.Length 6.3
## 350 versicolor Petal.Length 4.4
## 351 versicolor Sepal.Width 2.3
## 352 versicolor Petal.Width 1.3
## 353 versicolor Sepal.Length 5.6
## 354 versicolor Petal.Length 4.1
## 355 versicolor Sepal.Width 3.0
## 356 versicolor Petal.Width 1.3
## 357 versicolor Sepal.Length 5.5
## 358 versicolor Petal.Length 4.0
## 359 versicolor Sepal.Width 2.5
## 360 versicolor Petal.Width 1.3
## 361 versicolor Sepal.Length 5.5
## 362 versicolor Petal.Length 4.4
## 363 versicolor Sepal.Width 2.6
## 364 versicolor Petal.Width 1.2
## 365 versicolor Sepal.Length 6.1
## 366 versicolor Petal.Length 4.6
## 367 versicolor Sepal.Width 3.0
## 368 versicolor Petal.Width 1.4
## 369 versicolor Sepal.Length 5.8
## 370 versicolor Petal.Length 4.0
## 371 versicolor Sepal.Width 2.6
## 372 versicolor Petal.Width 1.2
## 373 versicolor Sepal.Length 5.0
## 374 versicolor Petal.Length 3.3
## 375 versicolor Sepal.Width 2.3
## 376 versicolor Petal.Width 1.0
## 377 versicolor Sepal.Length 5.6
## 378 versicolor Petal.Length 4.2
## 379 versicolor Sepal.Width 2.7
## 380 versicolor Petal.Width 1.3
## 381 versicolor Sepal.Length 5.7
## 382 versicolor Petal.Length 4.2
## 383 versicolor Sepal.Width 3.0
## 384 versicolor Petal.Width 1.2
## 385 versicolor Sepal.Length 5.7
## 386 versicolor Petal.Length 4.2
## 387 versicolor Sepal.Width 2.9
## 388 versicolor Petal.Width 1.3
## 389 versicolor Sepal.Length 6.2
## 390 versicolor Petal.Length 4.3
## 391 versicolor Sepal.Width 2.9
## 392 versicolor Petal.Width 1.3
## 393 versicolor Sepal.Length 5.1
## 394 versicolor Petal.Length 3.0
## 395 versicolor Sepal.Width 2.5
## 396 versicolor Petal.Width 1.1
## 397 versicolor Sepal.Length 5.7
## 398 versicolor Petal.Length 4.1
## 399 versicolor Sepal.Width 2.8
## 400 versicolor Petal.Width 1.3
## 401 virginica Sepal.Length 6.3
## 402 virginica Petal.Length 6.0
## 403 virginica Sepal.Width 3.3
## 404 virginica Petal.Width 2.5
## 405 virginica Sepal.Length 5.8
## 406 virginica Petal.Length 5.1
## 407 virginica Sepal.Width 2.7
## 408 virginica Petal.Width 1.9
## 409 virginica Sepal.Length 7.1
## 410 virginica Petal.Length 5.9
## 411 virginica Sepal.Width 3.0
## 412 virginica Petal.Width 2.1
## 413 virginica Sepal.Length 6.3
## 414 virginica Petal.Length 5.6
## 415 virginica Sepal.Width 2.9
## 416 virginica Petal.Width 1.8
## 417 virginica Sepal.Length 6.5
## 418 virginica Petal.Length 5.8
## 419 virginica Sepal.Width 3.0
## 420 virginica Petal.Width 2.2
## 421 virginica Sepal.Length 7.6
## 422 virginica Petal.Length 6.6
## 423 virginica Sepal.Width 3.0
## 424 virginica Petal.Width 2.1
## 425 virginica Sepal.Length 4.9
## 426 virginica Petal.Length 4.5
## 427 virginica Sepal.Width 2.5
## 428 virginica Petal.Width 1.7
## 429 virginica Sepal.Length 7.3
## 430 virginica Petal.Length 6.3
## 431 virginica Sepal.Width 2.9
## 432 virginica Petal.Width 1.8
## 433 virginica Sepal.Length 6.7
## 434 virginica Petal.Length 5.8
## 435 virginica Sepal.Width 2.5
## 436 virginica Petal.Width 1.8
## 437 virginica Sepal.Length 7.2
## 438 virginica Petal.Length 6.1
## 439 virginica Sepal.Width 3.6
## 440 virginica Petal.Width 2.5
## 441 virginica Sepal.Length 6.5
## 442 virginica Petal.Length 5.1
## 443 virginica Sepal.Width 3.2
## 444 virginica Petal.Width 2.0
## 445 virginica Sepal.Length 6.4
## 446 virginica Petal.Length 5.3
## 447 virginica Sepal.Width 2.7
## 448 virginica Petal.Width 1.9
## 449 virginica Sepal.Length 6.8
## 450 virginica Petal.Length 5.5
## 451 virginica Sepal.Width 3.0
## 452 virginica Petal.Width 2.1
## 453 virginica Sepal.Length 5.7
## 454 virginica Petal.Length 5.0
## 455 virginica Sepal.Width 2.5
## 456 virginica Petal.Width 2.0
## 457 virginica Sepal.Length 5.8
## 458 virginica Petal.Length 5.1
## 459 virginica Sepal.Width 2.8
## 460 virginica Petal.Width 2.4
## 461 virginica Sepal.Length 6.4
## 462 virginica Petal.Length 5.3
## 463 virginica Sepal.Width 3.2
## 464 virginica Petal.Width 2.3
## 465 virginica Sepal.Length 6.5
## 466 virginica Petal.Length 5.5
## 467 virginica Sepal.Width 3.0
## 468 virginica Petal.Width 1.8
## 469 virginica Sepal.Length 7.7
## 470 virginica Petal.Length 6.7
## 471 virginica Sepal.Width 3.8
## 472 virginica Petal.Width 2.2
## 473 virginica Sepal.Length 7.7
## 474 virginica Petal.Length 6.9
## 475 virginica Sepal.Width 2.6
## 476 virginica Petal.Width 2.3
## 477 virginica Sepal.Length 6.0
## 478 virginica Petal.Length 5.0
## 479 virginica Sepal.Width 2.2
## 480 virginica Petal.Width 1.5
## 481 virginica Sepal.Length 6.9
## 482 virginica Petal.Length 5.7
## 483 virginica Sepal.Width 3.2
## 484 virginica Petal.Width 2.3
## 485 virginica Sepal.Length 5.6
## 486 virginica Petal.Length 4.9
## 487 virginica Sepal.Width 2.8
## 488 virginica Petal.Width 2.0
## 489 virginica Sepal.Length 7.7
## 490 virginica Petal.Length 6.7
## 491 virginica Sepal.Width 2.8
## 492 virginica Petal.Width 2.0
## 493 virginica Sepal.Length 6.3
## 494 virginica Petal.Length 4.9
## 495 virginica Sepal.Width 2.7
## 496 virginica Petal.Width 1.8
## 497 virginica Sepal.Length 6.7
## 498 virginica Petal.Length 5.7
## 499 virginica Sepal.Width 3.3
## 500 virginica Petal.Width 2.1
## 501 virginica Sepal.Length 7.2
## 502 virginica Petal.Length 6.0
## 503 virginica Sepal.Width 3.2
## 504 virginica Petal.Width 1.8
## 505 virginica Sepal.Length 6.2
## 506 virginica Petal.Length 4.8
## 507 virginica Sepal.Width 2.8
## 508 virginica Petal.Width 1.8
## 509 virginica Sepal.Length 6.1
## 510 virginica Petal.Length 4.9
## 511 virginica Sepal.Width 3.0
## 512 virginica Petal.Width 1.8
## 513 virginica Sepal.Length 6.4
## 514 virginica Petal.Length 5.6
## 515 virginica Sepal.Width 2.8
## 516 virginica Petal.Width 2.1
## 517 virginica Sepal.Length 7.2
## 518 virginica Petal.Length 5.8
## 519 virginica Sepal.Width 3.0
## 520 virginica Petal.Width 1.6
## 521 virginica Sepal.Length 7.4
## 522 virginica Petal.Length 6.1
## 523 virginica Sepal.Width 2.8
## 524 virginica Petal.Width 1.9
## 525 virginica Sepal.Length 7.9
## 526 virginica Petal.Length 6.4
## 527 virginica Sepal.Width 3.8
## 528 virginica Petal.Width 2.0
## 529 virginica Sepal.Length 6.4
## 530 virginica Petal.Length 5.6
## 531 virginica Sepal.Width 2.8
## 532 virginica Petal.Width 2.2
## 533 virginica Sepal.Length 6.3
## 534 virginica Petal.Length 5.1
## 535 virginica Sepal.Width 2.8
## 536 virginica Petal.Width 1.5
## 537 virginica Sepal.Length 6.1
## 538 virginica Petal.Length 5.6
## 539 virginica Sepal.Width 2.6
## 540 virginica Petal.Width 1.4
## 541 virginica Sepal.Length 7.7
## 542 virginica Petal.Length 6.1
## 543 virginica Sepal.Width 3.0
## 544 virginica Petal.Width 2.3
## 545 virginica Sepal.Length 6.3
## 546 virginica Petal.Length 5.6
## 547 virginica Sepal.Width 3.4
## 548 virginica Petal.Width 2.4
## 549 virginica Sepal.Length 6.4
## 550 virginica Petal.Length 5.5
## 551 virginica Sepal.Width 3.1
## 552 virginica Petal.Width 1.8
## 553 virginica Sepal.Length 6.0
## 554 virginica Petal.Length 4.8
## 555 virginica Sepal.Width 3.0
## 556 virginica Petal.Width 1.8
## 557 virginica Sepal.Length 6.9
## 558 virginica Petal.Length 5.4
## 559 virginica Sepal.Width 3.1
## 560 virginica Petal.Width 2.1
## 561 virginica Sepal.Length 6.7
## 562 virginica Petal.Length 5.6
## 563 virginica Sepal.Width 3.1
## 564 virginica Petal.Width 2.4
## 565 virginica Sepal.Length 6.9
## 566 virginica Petal.Length 5.1
## 567 virginica Sepal.Width 3.1
## 568 virginica Petal.Width 2.3
## 569 virginica Sepal.Length 5.8
## 570 virginica Petal.Length 5.1
## 571 virginica Sepal.Width 2.7
## 572 virginica Petal.Width 1.9
## 573 virginica Sepal.Length 6.8
## 574 virginica Petal.Length 5.9
## 575 virginica Sepal.Width 3.2
## 576 virginica Petal.Width 2.3
## 577 virginica Sepal.Length 6.7
## 578 virginica Petal.Length 5.7
## 579 virginica Sepal.Width 3.3
## 580 virginica Petal.Width 2.5
## 581 virginica Sepal.Length 6.7
## 582 virginica Petal.Length 5.2
## 583 virginica Sepal.Width 3.0
## 584 virginica Petal.Width 2.3
## 585 virginica Sepal.Length 6.3
## 586 virginica Petal.Length 5.0
## 587 virginica Sepal.Width 2.5
## 588 virginica Petal.Width 1.9
## 589 virginica Sepal.Length 6.5
## 590 virginica Petal.Length 5.2
## 591 virginica Sepal.Width 3.0
## 592 virginica Petal.Width 2.0
## 593 virginica Sepal.Length 6.2
## 594 virginica Petal.Length 5.4
## 595 virginica Sepal.Width 3.4
## 596 virginica Petal.Width 2.3
## 597 virginica Sepal.Length 5.9
## 598 virginica Petal.Length 5.1
## 599 virginica Sepal.Width 3.0
## 600 virginica Petal.Width 1.8
Below we define a larger version of iris data, as a function of number
of rows N:
N <- 250
(row.id.vec <- 1+(seq(0,N-1) %% nrow(iris)))
## [1] 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28
## [29] 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56
## [57] 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71 72 73 74 75 76 77 78 79 80 81 82 83 84
## [85] 85 86 87 88 89 90 91 92 93 94 95 96 97 98 99 100 101 102 103 104 105 106 107 108 109 110 111 112
## [113] 113 114 115 116 117 118 119 120 121 122 123 124 125 126 127 128 129 130 131 132 133 134 135 136 137 138 139 140
## [141] 141 142 143 144 145 146 147 148 149 150 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18
## [169] 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46
## [197] 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71 72 73 74
## [225] 75 76 77 78 79 80 81 82 83 84 85 86 87 88 89 90 91 92 93 94 95 96 97 98 99 100
N.df <- iris[row.id.vec,]
(N.dt <- data.table(N.df))
## Sepal.Length Sepal.Width Petal.Length Petal.Width Species
## <num> <num> <num> <num> <fctr>
## 1: 5.1 3.5 1.4 0.2 setosa
## 2: 4.9 3.0 1.4 0.2 setosa
## 3: 4.7 3.2 1.3 0.2 setosa
## 4: 4.6 3.1 1.5 0.2 setosa
## 5: 5.0 3.6 1.4 0.2 setosa
## ---
## 246: 5.7 3.0 4.2 1.2 versicolor
## 247: 5.7 2.9 4.2 1.3 versicolor
## 248: 6.2 2.9 4.3 1.3 versicolor
## 249: 5.1 2.5 3.0 1.1 versicolor
## 250: 5.7 2.8 4.1 1.3 versicolor
Below we use atime to compare the performance of the reshape
functions:
a.res <- atime::atime(
N=2^seq(1,50),
setup={
(row.id.vec <- 1+(seq(0,N-1) %% nrow(iris)))
N.df <- iris[row.id.vec,]
(N.dt <- data.table(N.df))
polars_df <- polars::as_polars_df(N.df)
duckdb::dbWriteTable(con, "iris_table", N.df, overwrite=TRUE)
},
seconds.limit=0.1,
"duckdb\ndbWriteTable"=duckdb::dbWriteTable(con, "iris_table", N.df, overwrite=TRUE),
"duckdb\nUNPIVOT"=DBI::dbGetQuery(con, 'UNPIVOT iris_table ON "Sepal.Length", "Petal.Length", "Sepal.Width", "Petal.Width" INTO NAME part_dim VALUE cm'),
"polars\nas_polars_df"=polars::as_polars_df(N.df),
"polars\nunpivot"=polars_df$unpivot(index="Species", value_name="cm"),
"stats\nreshape"=suppressWarnings(stats::reshape(N.df, direction="long", varying=list(cols.to.reshape), v.names="cm")),
"data.table\nmelt"=melt(N.dt, measure.vars=cols.to.reshape, value.name="cm"),
"tidyr\npivot_longer"=tidyr::pivot_longer(N.df, cols.to.reshape, values_to = "cm"),
"collapse\npivot"=collapse::pivot(N.df, values=cols.to.reshape, names=list("variable", "cm")))
a.refs <- atime::references_best(a.res)
a.pred <- predict(a.refs)
plot(a.pred)+coord_cartesian(xlim=c(1e2,1e7))
## Le chargement a nécessité le package : directlabels
## Warning in ggplot2::scale_x_log10("N", breaks = meas[, 10^seq(ceiling(min(log10(N))), : log-10 transformation
## introduced infinite values.
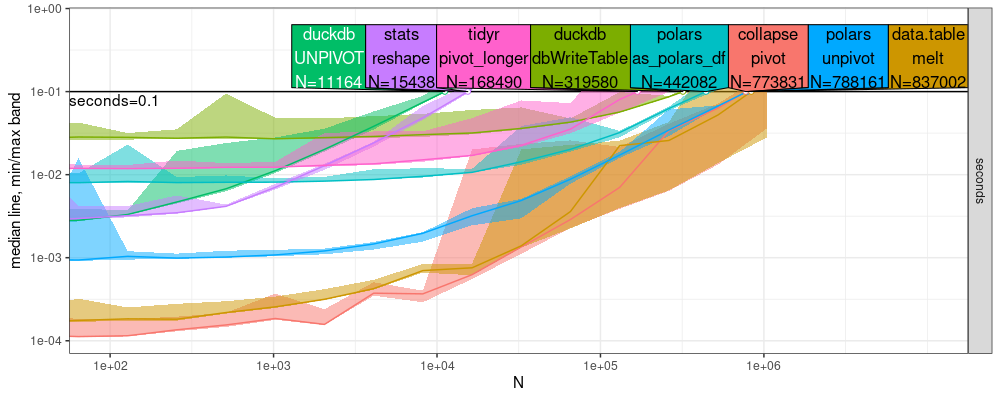
The result above shows that collapse is fastest, but data.table is
almost as fast (by small a constant factor).
Regex and multiple column support
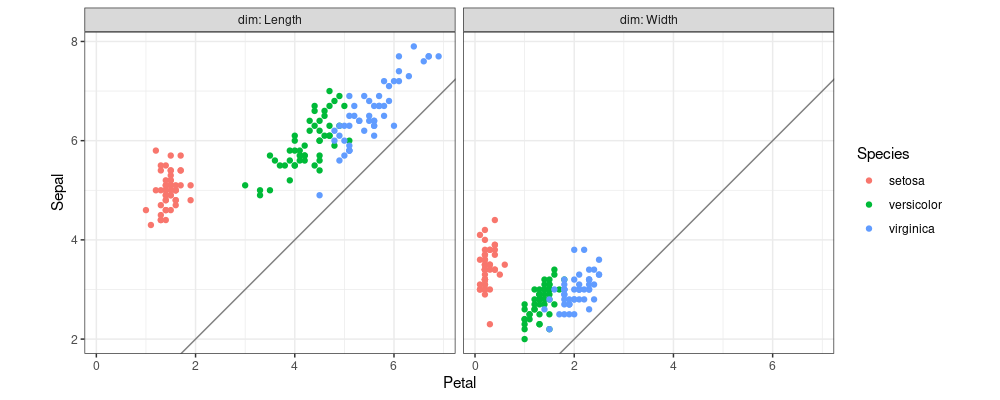
In my paper about the nc package, I proposed a new syntax for defining wide-to-tall reshape operations, using regular expressions (regex). The motivating example in that paper was a scatterplot to show that Sepals are larger than Petals, as we see in the code/plot below.
iris.parts <- nc::capture_melt_multiple(iris.dt, column=".*", "[.]", dim=".*")
ggplot()+
geom_point(aes(
Petal, Sepal, color=Species),
data=iris.parts)+
facet_grid(. ~ dim, labeller=label_both)+
coord_equal()+
theme_bw()+
geom_abline(slope=1, intercept=0, color="grey50")
Other ways to do that reshape operation are shown in the code below:
stats::reshape(two.iris.wide, cols.to.reshape, direction="long", timevar="dim", sep=".")
## Species dim Sepal Petal id
## 1.Length setosa Length 5.1 1.4 1
## 2.Length virginica Length 5.9 5.1 2
## 1.Width setosa Width 3.5 0.2 1
## 2.Width virginica Width 3.0 1.8 2
melt(data.table(two.iris.wide), measure.vars=measure(value.name, part, pattern="(.*)[.](.*)"))
## Species part Sepal Petal
## <fctr> <char> <num> <num>
## 1: setosa Length 5.1 1.4
## 2: virginica Length 5.9 5.1
## 3: setosa Width 3.5 0.2
## 4: virginica Width 3.0 1.8
tidyr::pivot_longer(two.iris.wide, cols.to.reshape, names_pattern = "(.*)[.](.*)", names_to = c(".value","part"))
## # A tibble: 4 × 4
## Species part Sepal Petal
## <fct> <chr> <dbl> <dbl>
## 1 setosa Length 5.1 1.4
## 2 setosa Width 3.5 0.2
## 3 virginica Length 5.9 5.1
## 4 virginica Width 3 1.8
However ?collapse::pivot explains that it “currently does not
support concurrently melting/pivoting longer to multiple columns.”
And unpivot does not support multiple output columns either.
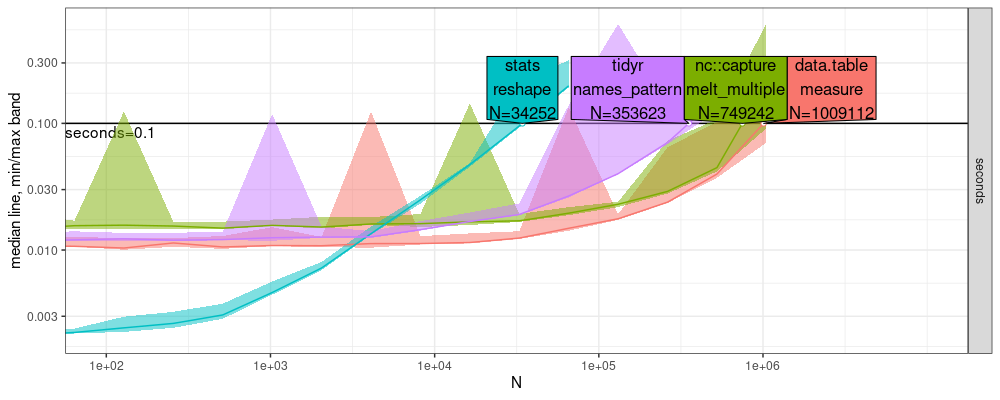
Below we compare the performance of the methods that do support multiple output columns:
r.res <- atime::atime(
N=2^seq(1,50),
setup={
(row.id.vec <- 1+(seq(0,N-1) %% nrow(iris)))
N.df <- iris[row.id.vec,]
(N.dt <- data.table(N.df))
},
seconds.limit=0.1,
"nc::capture\nmelt_multiple"=nc::capture_melt_multiple(N.dt, column=".*", "[.]", dim=".*"),
"stats\nreshape"=stats::reshape(N.df, cols.to.reshape, direction="long", timevar="dim", sep="."),
"data.table\nmeasure"=melt(N.dt, measure.vars=measure(value.name, part, pattern="(.*)[.](.*)")),
"tidyr\nnames_pattern"=tidyr::pivot_longer(N.df, cols.to.reshape, names_pattern = "(.*)[.](.*)", names_to = c(".value","part")))
r.refs <- atime::references_best(r.res)
r.pred <- predict(r.refs)
plot(r.pred)+coord_cartesian(xlim=c(1e2,1e7))
## Warning in ggplot2::scale_x_log10("N", breaks = meas[, 10^seq(ceiling(min(log10(N))), : log-10 transformation
## introduced infinite values.
Reshape wider
Above we did a wide to tall reshape. The inverse of that operation is a tall to wide reshape,
matrix(
two.iris.tall$cm, nrow(two.iris.wide), length(cols.to.reshape),
dimnames = list(
two.iris.tall$row.name[1:nrow(two.iris.wide)],
cols.to.reshape))
## Sepal.Length Sepal.Width Petal.Length Petal.Width
## 1 5.1 3.5 1.4 0.2
## 150 5.9 3.0 5.1 1.8
More generically to do that, we would need to consider missing entries, as in the code below.
reshape_wider <- function(DF){
dimnames <- list()
other.names <- setdiff(
names(DF),
c("orig.row.i", "orig.row.name", "orig.col.i", "orig.col.name", "value"))
for(dim.type in c('row','col')){
f <- function(suffix)paste0('orig.',dim.type,'.',suffix)
u <- unique(DF[, c(
f(c("i","name")),
if(dim.type=='row')other.names
)])
s <- u[order(u[,f("i")]), ]
dimnames[[dim.type]] <- s[, f("name")]
if(dim.type=='row')other.df <- s[,other.names,drop=FALSE]
}
mat <- matrix(
NA_real_, length(dimnames$row), length(dimnames$col),
dimnames = dimnames)
mat[cbind(DF$orig.row.i, DF$orig.col.i)] <- DF$value
data.frame(mat, other.df)
}
(missing.one <- reshape_taller(two.iris.wide,1:4)[-1,])
## orig.row.i orig.row.name orig.col.i orig.col.name Species value
## 2 2 150 1 Sepal.Length virginica 5.9
## 3 1 1 2 Sepal.Width setosa 3.5
## 4 2 150 2 Sepal.Width virginica 3.0
## 5 1 1 3 Petal.Length setosa 1.4
## 6 2 150 3 Petal.Length virginica 5.1
## 7 1 1 4 Petal.Width setosa 0.2
## 8 2 150 4 Petal.Width virginica 1.8
reshape_wider(missing.one)
## Sepal.Length Sepal.Width Petal.Length Petal.Width Species
## 1 NA 3.5 1.4 0.2 setosa
## 150 5.9 3.0 5.1 1.8 virginica
Other functions for doing that below:
library(data.table)
N.tall.df <- reshape_taller(N.df,1:4)
str(reshape_wider(N.tall.df))
## 'data.frame': 250 obs. of 5 variables:
## $ Sepal.Length: num 5.1 4.9 4.7 4.6 5 5.4 4.6 5 4.4 4.9 ...
## $ Sepal.Width : num 3.5 3 3.2 3.1 3.6 3.9 3.4 3.4 2.9 3.1 ...
## $ Petal.Length: num 1.4 1.4 1.3 1.5 1.4 1.7 1.4 1.5 1.4 1.5 ...
## $ Petal.Width : num 0.2 0.2 0.2 0.2 0.2 0.4 0.3 0.2 0.2 0.1 ...
## $ Species : Factor w/ 3 levels "setosa","versicolor",..: 1 1 1 1 1 1 1 1 1 1 ...
N.tall.dt <- data.table(N.tall.df)
str(dcast(N.tall.dt, orig.row.i + Species ~ orig.col.name, value.var = "value"))
## Classes 'data.table' and 'data.frame': 250 obs. of 6 variables:
## $ orig.row.i : int 1 2 3 4 5 6 7 8 9 10 ...
## $ Species : Factor w/ 3 levels "setosa","versicolor",..: 1 1 1 1 1 1 1 1 1 1 ...
## $ Petal.Length: num 1.4 1.4 1.3 1.5 1.4 1.7 1.4 1.5 1.4 1.5 ...
## $ Petal.Width : num 0.2 0.2 0.2 0.2 0.2 0.4 0.3 0.2 0.2 0.1 ...
## $ Sepal.Length: num 5.1 4.9 4.7 4.6 5 5.4 4.6 5 4.4 4.9 ...
## $ Sepal.Width : num 3.5 3 3.2 3.1 3.6 3.9 3.4 3.4 2.9 3.1 ...
## - attr(*, ".internal.selfref")=<externalptr>
## - attr(*, "sorted")= chr [1:2] "orig.row.i" "Species"
str(stats::reshape(N.tall.df, direction = "wide", idvar=c("orig.row.i","Species"), timevar="orig.col.name", v.names="value"))
## Warning in reshapeWide(data, idvar = idvar, timevar = timevar, varying = varying, : certaines variables constantes
## (orig.col.i) varient
## 'data.frame': 250 obs. of 8 variables:
## $ orig.row.i : int 1 2 3 4 5 6 7 8 9 10 ...
## $ orig.row.name : chr "1" "2" "3" "4" ...
## $ orig.col.i : int 1 1 1 1 1 1 1 1 1 1 ...
## $ Species : Factor w/ 3 levels "setosa","versicolor",..: 1 1 1 1 1 1 1 1 1 1 ...
## $ value.Sepal.Length: num 5.1 4.9 4.7 4.6 5 5.4 4.6 5 4.4 4.9 ...
## $ value.Sepal.Width : num 3.5 3 3.2 3.1 3.6 3.9 3.4 3.4 2.9 3.1 ...
## $ value.Petal.Length: num 1.4 1.4 1.3 1.5 1.4 1.7 1.4 1.5 1.4 1.5 ...
## $ value.Petal.Width : num 0.2 0.2 0.2 0.2 0.2 0.4 0.3 0.2 0.2 0.1 ...
## - attr(*, "reshapeWide")=List of 5
## ..$ v.names: chr "value"
## ..$ timevar: chr "orig.col.name"
## ..$ idvar : chr [1:2] "orig.row.i" "Species"
## ..$ times : chr [1:4] "Sepal.Length" "Sepal.Width" "Petal.Length" "Petal.Width"
## ..$ varying: chr [1, 1:4] "value.Sepal.Length" "value.Sepal.Width" "value.Petal.Length" "value.Petal.Width"
str(tidyr::pivot_wider(N.tall.df, names_from=orig.col.name, values_from=value, id_cols=c(orig.row.i,Species)))
## tibble [250 × 6] (S3: tbl_df/tbl/data.frame)
## $ orig.row.i : int [1:250] 1 2 3 4 5 6 7 8 9 10 ...
## $ Species : Factor w/ 3 levels "setosa","versicolor",..: 1 1 1 1 1 1 1 1 1 1 ...
## $ Sepal.Length: num [1:250] 5.1 4.9 4.7 4.6 5 5.4 4.6 5 4.4 4.9 ...
## $ Sepal.Width : num [1:250] 3.5 3 3.2 3.1 3.6 3.9 3.4 3.4 2.9 3.1 ...
## $ Petal.Length: num [1:250] 1.4 1.4 1.3 1.5 1.4 1.7 1.4 1.5 1.4 1.5 ...
## $ Petal.Width : num [1:250] 0.2 0.2 0.2 0.2 0.2 0.4 0.3 0.2 0.2 0.1 ...
str(collapse::pivot(N.tall.df, how="w", ids=c("orig.row.i","Species"), values="value", names="orig.col.name"))
## 'data.frame': 250 obs. of 6 variables:
## $ orig.row.i : int 1 2 3 4 5 6 7 8 9 10 ...
## $ Species : Factor w/ 3 levels "setosa","versicolor",..: 1 1 1 1 1 1 1 1 1 1 ...
## $ Sepal.Length: num 5.1 4.9 4.7 4.6 5 5.4 4.6 5 4.4 4.9 ...
## $ Sepal.Width : num 3.5 3 3.2 3.1 3.6 3.9 3.4 3.4 2.9 3.1 ...
## $ Petal.Length: num 1.4 1.4 1.3 1.5 1.4 1.7 1.4 1.5 1.4 1.5 ...
## $ Petal.Width : num 0.2 0.2 0.2 0.2 0.2 0.4 0.3 0.2 0.2 0.1 ...
polars::as_polars_df(N.tall.df)$pivot(on="orig.col.name", index=c("orig.row.i","Species"), values="value")
<div><style>
.dataframe > thead > tr > th,
.dataframe > tbody > tr > td {
text-align: right;
white-space: pre-wrap;
}
</style>
<small>shape: (250, 6)</small><table border="1" class="dataframe"><thead><tr><th>orig.row.i</th><th>Species</th><th>Sepal.Length</th><th>Sepal.Width</th><th>Petal.Length</th><th>Petal.Width</th></tr><tr><td>i32</td><td>cat</td><td>f64</td><td>f64</td><td>f64</td><td>f64</td></tr></thead><tbody><tr><td>1</td><td>"setosa"</td><td>5.1</td><td>3.5</td><td>1.4</td><td>0.2</td></tr><tr><td>2</td><td>"setosa"</td><td>4.9</td><td>3.0</td><td>1.4</td><td>0.2</td></tr><tr><td>3</td><td>"setosa"</td><td>4.7</td><td>3.2</td><td>1.3</td><td>0.2</td></tr><tr><td>4</td><td>"setosa"</td><td>4.6</td><td>3.1</td><td>1.5</td><td>0.2</td></tr><tr><td>5</td><td>"setosa"</td><td>5.0</td><td>3.6</td><td>1.4</td><td>0.2</td></tr><tr><td>…</td><td>…</td><td>…</td><td>…</td><td>…</td><td>…</td></tr><tr><td>246</td><td>"versicolor"</td><td>5.7</td><td>3.0</td><td>4.2</td><td>1.2</td></tr><tr><td>247</td><td>"versicolor"</td><td>5.7</td><td>2.9</td><td>4.2</td><td>1.3</td></tr><tr><td>248</td><td>"versicolor"</td><td>6.2</td><td>2.9</td><td>4.3</td><td>1.3</td></tr><tr><td>249</td><td>"versicolor"</td><td>5.1</td><td>2.5</td><td>3.0</td><td>1.1</td></tr><tr><td>250</td><td>"versicolor"</td><td>5.7</td><td>2.8</td><td>4.1</td><td>1.3</td></tr></tbody></table></div>
duckdb::dbWriteTable(con, "iris_tall", N.tall.df, overwrite=TRUE)
DBI::dbGetQuery(con, 'PIVOT iris_tall ON "orig.col.name" USING sum(value) GROUP BY "orig.row.i", "orig.row.name" ORDER BY "orig.row.i"')
## orig.row.i orig.row.name Petal.Length Petal.Width Sepal.Length Sepal.Width
## 1 1 1 1.4 0.2 5.1 3.5
## 2 2 2 1.4 0.2 4.9 3.0
## 3 3 3 1.3 0.2 4.7 3.2
## 4 4 4 1.5 0.2 4.6 3.1
## 5 5 5 1.4 0.2 5.0 3.6
## 6 6 6 1.7 0.4 5.4 3.9
## 7 7 7 1.4 0.3 4.6 3.4
## 8 8 8 1.5 0.2 5.0 3.4
## 9 9 9 1.4 0.2 4.4 2.9
## 10 10 10 1.5 0.1 4.9 3.1
## 11 11 11 1.5 0.2 5.4 3.7
## 12 12 12 1.6 0.2 4.8 3.4
## 13 13 13 1.4 0.1 4.8 3.0
## 14 14 14 1.1 0.1 4.3 3.0
## 15 15 15 1.2 0.2 5.8 4.0
## 16 16 16 1.5 0.4 5.7 4.4
## 17 17 17 1.3 0.4 5.4 3.9
## 18 18 18 1.4 0.3 5.1 3.5
## 19 19 19 1.7 0.3 5.7 3.8
## 20 20 20 1.5 0.3 5.1 3.8
## 21 21 21 1.7 0.2 5.4 3.4
## 22 22 22 1.5 0.4 5.1 3.7
## 23 23 23 1.0 0.2 4.6 3.6
## 24 24 24 1.7 0.5 5.1 3.3
## 25 25 25 1.9 0.2 4.8 3.4
## 26 26 26 1.6 0.2 5.0 3.0
## 27 27 27 1.6 0.4 5.0 3.4
## 28 28 28 1.5 0.2 5.2 3.5
## 29 29 29 1.4 0.2 5.2 3.4
## 30 30 30 1.6 0.2 4.7 3.2
## 31 31 31 1.6 0.2 4.8 3.1
## 32 32 32 1.5 0.4 5.4 3.4
## 33 33 33 1.5 0.1 5.2 4.1
## 34 34 34 1.4 0.2 5.5 4.2
## 35 35 35 1.5 0.2 4.9 3.1
## 36 36 36 1.2 0.2 5.0 3.2
## 37 37 37 1.3 0.2 5.5 3.5
## 38 38 38 1.4 0.1 4.9 3.6
## 39 39 39 1.3 0.2 4.4 3.0
## 40 40 40 1.5 0.2 5.1 3.4
## 41 41 41 1.3 0.3 5.0 3.5
## 42 42 42 1.3 0.3 4.5 2.3
## 43 43 43 1.3 0.2 4.4 3.2
## 44 44 44 1.6 0.6 5.0 3.5
## 45 45 45 1.9 0.4 5.1 3.8
## 46 46 46 1.4 0.3 4.8 3.0
## 47 47 47 1.6 0.2 5.1 3.8
## 48 48 48 1.4 0.2 4.6 3.2
## 49 49 49 1.5 0.2 5.3 3.7
## 50 50 50 1.4 0.2 5.0 3.3
## 51 51 51 4.7 1.4 7.0 3.2
## 52 52 52 4.5 1.5 6.4 3.2
## 53 53 53 4.9 1.5 6.9 3.1
## 54 54 54 4.0 1.3 5.5 2.3
## 55 55 55 4.6 1.5 6.5 2.8
## 56 56 56 4.5 1.3 5.7 2.8
## 57 57 57 4.7 1.6 6.3 3.3
## 58 58 58 3.3 1.0 4.9 2.4
## 59 59 59 4.6 1.3 6.6 2.9
## 60 60 60 3.9 1.4 5.2 2.7
## 61 61 61 3.5 1.0 5.0 2.0
## 62 62 62 4.2 1.5 5.9 3.0
## 63 63 63 4.0 1.0 6.0 2.2
## 64 64 64 4.7 1.4 6.1 2.9
## 65 65 65 3.6 1.3 5.6 2.9
## 66 66 66 4.4 1.4 6.7 3.1
## 67 67 67 4.5 1.5 5.6 3.0
## 68 68 68 4.1 1.0 5.8 2.7
## 69 69 69 4.5 1.5 6.2 2.2
## 70 70 70 3.9 1.1 5.6 2.5
## 71 71 71 4.8 1.8 5.9 3.2
## 72 72 72 4.0 1.3 6.1 2.8
## 73 73 73 4.9 1.5 6.3 2.5
## 74 74 74 4.7 1.2 6.1 2.8
## 75 75 75 4.3 1.3 6.4 2.9
## 76 76 76 4.4 1.4 6.6 3.0
## 77 77 77 4.8 1.4 6.8 2.8
## 78 78 78 5.0 1.7 6.7 3.0
## 79 79 79 4.5 1.5 6.0 2.9
## 80 80 80 3.5 1.0 5.7 2.6
## 81 81 81 3.8 1.1 5.5 2.4
## 82 82 82 3.7 1.0 5.5 2.4
## 83 83 83 3.9 1.2 5.8 2.7
## 84 84 84 5.1 1.6 6.0 2.7
## 85 85 85 4.5 1.5 5.4 3.0
## 86 86 86 4.5 1.6 6.0 3.4
## 87 87 87 4.7 1.5 6.7 3.1
## 88 88 88 4.4 1.3 6.3 2.3
## 89 89 89 4.1 1.3 5.6 3.0
## 90 90 90 4.0 1.3 5.5 2.5
## 91 91 91 4.4 1.2 5.5 2.6
## 92 92 92 4.6 1.4 6.1 3.0
## 93 93 93 4.0 1.2 5.8 2.6
## 94 94 94 3.3 1.0 5.0 2.3
## 95 95 95 4.2 1.3 5.6 2.7
## 96 96 96 4.2 1.2 5.7 3.0
## 97 97 97 4.2 1.3 5.7 2.9
## 98 98 98 4.3 1.3 6.2 2.9
## 99 99 99 3.0 1.1 5.1 2.5
## 100 100 100 4.1 1.3 5.7 2.8
## 101 101 101 6.0 2.5 6.3 3.3
## 102 102 102 5.1 1.9 5.8 2.7
## 103 103 103 5.9 2.1 7.1 3.0
## 104 104 104 5.6 1.8 6.3 2.9
## 105 105 105 5.8 2.2 6.5 3.0
## 106 106 106 6.6 2.1 7.6 3.0
## 107 107 107 4.5 1.7 4.9 2.5
## 108 108 108 6.3 1.8 7.3 2.9
## 109 109 109 5.8 1.8 6.7 2.5
## 110 110 110 6.1 2.5 7.2 3.6
## 111 111 111 5.1 2.0 6.5 3.2
## 112 112 112 5.3 1.9 6.4 2.7
## 113 113 113 5.5 2.1 6.8 3.0
## 114 114 114 5.0 2.0 5.7 2.5
## 115 115 115 5.1 2.4 5.8 2.8
## 116 116 116 5.3 2.3 6.4 3.2
## 117 117 117 5.5 1.8 6.5 3.0
## 118 118 118 6.7 2.2 7.7 3.8
## 119 119 119 6.9 2.3 7.7 2.6
## 120 120 120 5.0 1.5 6.0 2.2
## 121 121 121 5.7 2.3 6.9 3.2
## 122 122 122 4.9 2.0 5.6 2.8
## 123 123 123 6.7 2.0 7.7 2.8
## 124 124 124 4.9 1.8 6.3 2.7
## 125 125 125 5.7 2.1 6.7 3.3
## 126 126 126 6.0 1.8 7.2 3.2
## 127 127 127 4.8 1.8 6.2 2.8
## 128 128 128 4.9 1.8 6.1 3.0
## 129 129 129 5.6 2.1 6.4 2.8
## 130 130 130 5.8 1.6 7.2 3.0
## 131 131 131 6.1 1.9 7.4 2.8
## 132 132 132 6.4 2.0 7.9 3.8
## 133 133 133 5.6 2.2 6.4 2.8
## 134 134 134 5.1 1.5 6.3 2.8
## 135 135 135 5.6 1.4 6.1 2.6
## 136 136 136 6.1 2.3 7.7 3.0
## 137 137 137 5.6 2.4 6.3 3.4
## 138 138 138 5.5 1.8 6.4 3.1
## 139 139 139 4.8 1.8 6.0 3.0
## 140 140 140 5.4 2.1 6.9 3.1
## 141 141 141 5.6 2.4 6.7 3.1
## 142 142 142 5.1 2.3 6.9 3.1
## 143 143 143 5.1 1.9 5.8 2.7
## 144 144 144 5.9 2.3 6.8 3.2
## 145 145 145 5.7 2.5 6.7 3.3
## 146 146 146 5.2 2.3 6.7 3.0
## 147 147 147 5.0 1.9 6.3 2.5
## 148 148 148 5.2 2.0 6.5 3.0
## 149 149 149 5.4 2.3 6.2 3.4
## 150 150 150 5.1 1.8 5.9 3.0
## 151 151 1.1 1.4 0.2 5.1 3.5
## 152 152 2.1 1.4 0.2 4.9 3.0
## 153 153 3.1 1.3 0.2 4.7 3.2
## 154 154 4.1 1.5 0.2 4.6 3.1
## 155 155 5.1 1.4 0.2 5.0 3.6
## 156 156 6.1 1.7 0.4 5.4 3.9
## 157 157 7.1 1.4 0.3 4.6 3.4
## 158 158 8.1 1.5 0.2 5.0 3.4
## 159 159 9.1 1.4 0.2 4.4 2.9
## 160 160 10.1 1.5 0.1 4.9 3.1
## 161 161 11.1 1.5 0.2 5.4 3.7
## 162 162 12.1 1.6 0.2 4.8 3.4
## 163 163 13.1 1.4 0.1 4.8 3.0
## 164 164 14.1 1.1 0.1 4.3 3.0
## 165 165 15.1 1.2 0.2 5.8 4.0
## 166 166 16.1 1.5 0.4 5.7 4.4
## 167 167 17.1 1.3 0.4 5.4 3.9
## 168 168 18.1 1.4 0.3 5.1 3.5
## 169 169 19.1 1.7 0.3 5.7 3.8
## 170 170 20.1 1.5 0.3 5.1 3.8
## 171 171 21.1 1.7 0.2 5.4 3.4
## 172 172 22.1 1.5 0.4 5.1 3.7
## 173 173 23.1 1.0 0.2 4.6 3.6
## 174 174 24.1 1.7 0.5 5.1 3.3
## 175 175 25.1 1.9 0.2 4.8 3.4
## 176 176 26.1 1.6 0.2 5.0 3.0
## 177 177 27.1 1.6 0.4 5.0 3.4
## 178 178 28.1 1.5 0.2 5.2 3.5
## 179 179 29.1 1.4 0.2 5.2 3.4
## 180 180 30.1 1.6 0.2 4.7 3.2
## 181 181 31.1 1.6 0.2 4.8 3.1
## 182 182 32.1 1.5 0.4 5.4 3.4
## 183 183 33.1 1.5 0.1 5.2 4.1
## 184 184 34.1 1.4 0.2 5.5 4.2
## 185 185 35.1 1.5 0.2 4.9 3.1
## 186 186 36.1 1.2 0.2 5.0 3.2
## 187 187 37.1 1.3 0.2 5.5 3.5
## 188 188 38.1 1.4 0.1 4.9 3.6
## 189 189 39.1 1.3 0.2 4.4 3.0
## 190 190 40.1 1.5 0.2 5.1 3.4
## 191 191 41.1 1.3 0.3 5.0 3.5
## 192 192 42.1 1.3 0.3 4.5 2.3
## 193 193 43.1 1.3 0.2 4.4 3.2
## 194 194 44.1 1.6 0.6 5.0 3.5
## 195 195 45.1 1.9 0.4 5.1 3.8
## 196 196 46.1 1.4 0.3 4.8 3.0
## 197 197 47.1 1.6 0.2 5.1 3.8
## 198 198 48.1 1.4 0.2 4.6 3.2
## 199 199 49.1 1.5 0.2 5.3 3.7
## 200 200 50.1 1.4 0.2 5.0 3.3
## 201 201 51.1 4.7 1.4 7.0 3.2
## 202 202 52.1 4.5 1.5 6.4 3.2
## 203 203 53.1 4.9 1.5 6.9 3.1
## 204 204 54.1 4.0 1.3 5.5 2.3
## 205 205 55.1 4.6 1.5 6.5 2.8
## 206 206 56.1 4.5 1.3 5.7 2.8
## 207 207 57.1 4.7 1.6 6.3 3.3
## 208 208 58.1 3.3 1.0 4.9 2.4
## 209 209 59.1 4.6 1.3 6.6 2.9
## 210 210 60.1 3.9 1.4 5.2 2.7
## 211 211 61.1 3.5 1.0 5.0 2.0
## 212 212 62.1 4.2 1.5 5.9 3.0
## 213 213 63.1 4.0 1.0 6.0 2.2
## 214 214 64.1 4.7 1.4 6.1 2.9
## 215 215 65.1 3.6 1.3 5.6 2.9
## 216 216 66.1 4.4 1.4 6.7 3.1
## 217 217 67.1 4.5 1.5 5.6 3.0
## 218 218 68.1 4.1 1.0 5.8 2.7
## 219 219 69.1 4.5 1.5 6.2 2.2
## 220 220 70.1 3.9 1.1 5.6 2.5
## 221 221 71.1 4.8 1.8 5.9 3.2
## 222 222 72.1 4.0 1.3 6.1 2.8
## 223 223 73.1 4.9 1.5 6.3 2.5
## 224 224 74.1 4.7 1.2 6.1 2.8
## 225 225 75.1 4.3 1.3 6.4 2.9
## 226 226 76.1 4.4 1.4 6.6 3.0
## 227 227 77.1 4.8 1.4 6.8 2.8
## 228 228 78.1 5.0 1.7 6.7 3.0
## 229 229 79.1 4.5 1.5 6.0 2.9
## 230 230 80.1 3.5 1.0 5.7 2.6
## 231 231 81.1 3.8 1.1 5.5 2.4
## 232 232 82.1 3.7 1.0 5.5 2.4
## 233 233 83.1 3.9 1.2 5.8 2.7
## 234 234 84.1 5.1 1.6 6.0 2.7
## 235 235 85.1 4.5 1.5 5.4 3.0
## 236 236 86.1 4.5 1.6 6.0 3.4
## 237 237 87.1 4.7 1.5 6.7 3.1
## 238 238 88.1 4.4 1.3 6.3 2.3
## 239 239 89.1 4.1 1.3 5.6 3.0
## 240 240 90.1 4.0 1.3 5.5 2.5
## 241 241 91.1 4.4 1.2 5.5 2.6
## 242 242 92.1 4.6 1.4 6.1 3.0
## 243 243 93.1 4.0 1.2 5.8 2.6
## 244 244 94.1 3.3 1.0 5.0 2.3
## 245 245 95.1 4.2 1.3 5.6 2.7
## 246 246 96.1 4.2 1.2 5.7 3.0
## 247 247 97.1 4.2 1.3 5.7 2.9
## 248 248 98.1 4.3 1.3 6.2 2.9
## 249 249 99.1 3.0 1.1 5.1 2.5
## 250 250 100.1 4.1 1.3 5.7 2.8
In all of the code examples above there are a few common elements
- ID: used to identify unique output rows must be specified,
- Name: used for column names of output,
- Value: used to fill in elements of output.
| function | ID | Name | Value |
|---|---|---|---|
data.table::dcast |
LHS of formula | RHS of formula | value.var |
stats::reshape |
idvar |
timevar |
v.names |
tidyr::pivot_wider |
id_cols |
names_from |
values_from |
collapse::pivot |
ids |
names |
values |
polars $pivot |
index |
on |
values |
Timings below
w.res <- atime::atime(
N=2^seq(1,50),
setup={
(row.id.vec <- 1+(seq(0,N-1) %% nrow(iris)))
N.df <- iris[row.id.vec,]
N.tall.df <- reshape_taller(N.df,1:4)
N.tall.dt <- data.table(N.tall.df)
polars.df <- polars::as_polars_df(N.tall.df)
duckdb::dbWriteTable(con, "iris_tall", N.tall.df, overwrite=TRUE)
},
seconds.limit=0.1,
"duckdb\ndbWriteTable"=duckdb::dbWriteTable(con, "iris_tall", N.tall.df, overwrite=TRUE),
"duckdb\nPIVOT"=DBI::dbGetQuery(con, 'PIVOT iris_tall ON "orig.col.name" USING sum(value) GROUP BY "orig.row.i", "orig.row.name" ORDER BY "orig.row.i"'),
"polars\nas_polars_df"=polars::as_polars_df(N.tall.df),
"polars\npivot"=polars.df$pivot(on="orig.col.name", index=c("orig.row.i","Species"), values="value"),
"data.table\ndcast"=dcast(N.tall.dt, orig.row.i ~ orig.col.name, value.var = "value"),
"stats\nreshape"=suppressWarnings(stats::reshape(N.tall.df, direction = "wide", idvar=c("orig.row.i","Species"), timevar="orig.col.name", v.names="value")),
"tidyr\npivot_wider"=tidyr::pivot_wider(N.tall.df, names_from=orig.col.name, values_from=value, id_cols=orig.row.i),
"collapse\npivot"=collapse::pivot(N.tall.df, how="w", ids="orig.row.i", values="value", names="orig.col.name"))
w.refs <- atime::references_best(w.res)
w.pred <- predict(w.refs)
plot(w.pred)+coord_cartesian(xlim=c(NA,1e7))
## Warning in ggplot2::scale_x_log10("N", breaks = meas[, 10^seq(ceiling(min(log10(N))), : log-10 transformation
## introduced infinite values.
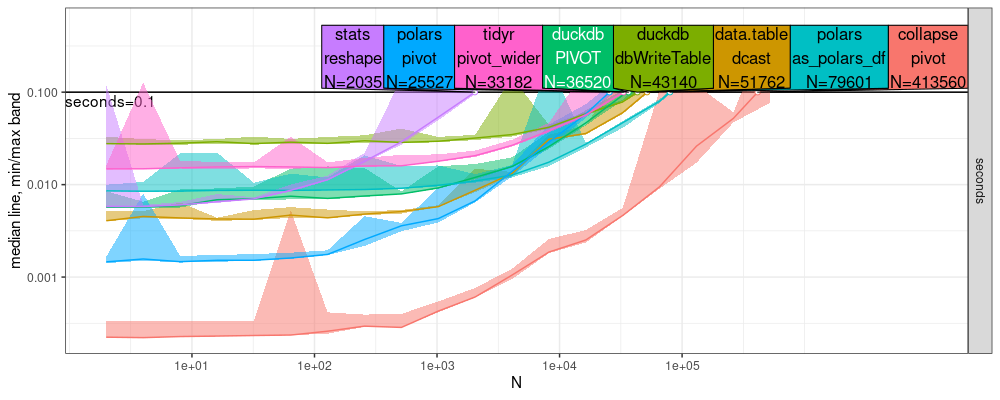
The comparison above shows that collapse is actually a bit faster
than data.table, for this tall to wide reshape operation which
involved just copying data (no summarization). But close inspection of
the log-log plot above shows different slopes for the different lines,
which suggests that they have different asymptotic complexity
classes. To estimate them, we use the code below,
w.refs$plot.references <- w.refs$ref[fun.name %in% c("N","N log N")]
plot(w.refs)
## Warning in ggplot2::scale_y_log10(""): log-10 transformation introduced infinite values.
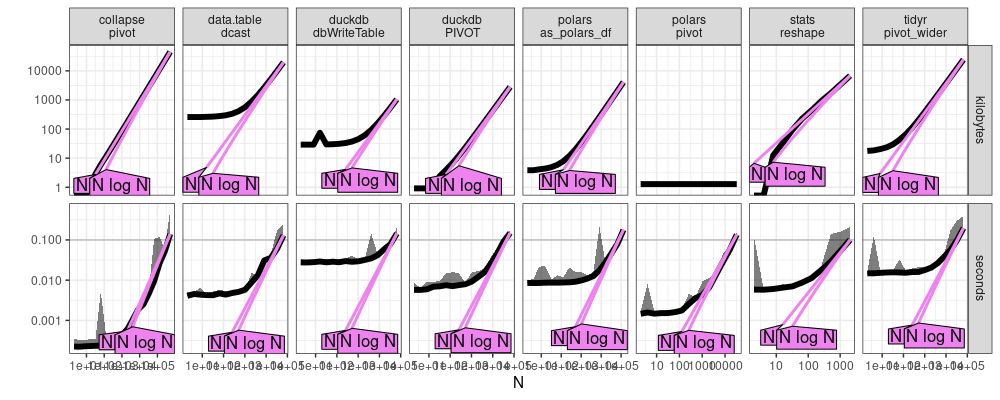
The plot above suggests that all methods have the same linear asymptotic memory usage, O(N), where N is the number of input rows.
data.table::dcastandstats::reshapeappear to be clearly linear time,O(N).collapse::pivotandtidyr::pivot_widermay be log-linear,O(N log N).
Reshape wider with summarization
Sometimes we want to do a reshape wider operation with summarization
functions. For example, in the code below, we use mean for every orig.col.name and Species:
dcast(N.tall.dt, orig.col.name + Species ~ ., mean)
## Key: <orig.col.name, Species>
## orig.col.name Species .
## <char> <fctr> <num>
## 1: Petal.Length setosa 1.462
## 2: Petal.Length versicolor 4.260
## 3: Petal.Length virginica 5.552
## 4: Petal.Width setosa 0.246
## 5: Petal.Width versicolor 1.326
## 6: Petal.Width virginica 2.026
## 7: Sepal.Length setosa 5.006
## 8: Sepal.Length versicolor 5.936
## 9: Sepal.Length virginica 6.588
## 10: Sepal.Width setosa 3.428
## 11: Sepal.Width versicolor 2.770
## 12: Sepal.Width virginica 2.974
stats::aggregate(N.tall.df[,"value",drop=FALSE], by=with(N.tall.df, list(orig.col.name=orig.col.name,Species=Species)), FUN=mean)
## orig.col.name Species value
## 1 Petal.Length setosa 1.462
## 2 Petal.Width setosa 0.246
## 3 Sepal.Length setosa 5.006
## 4 Sepal.Width setosa 3.428
## 5 Petal.Length versicolor 4.260
## 6 Petal.Width versicolor 1.326
## 7 Sepal.Length versicolor 5.936
## 8 Sepal.Width versicolor 2.770
## 9 Petal.Length virginica 5.552
## 10 Petal.Width virginica 2.026
## 11 Sepal.Length virginica 6.588
## 12 Sepal.Width virginica 2.974
N.tall.df$name <- "foo"
tidyr::pivot_wider(N.tall.df, names_from=name, values_from=value, id_cols=c(orig.col.name,Species), values_fn=mean)
## # A tibble: 12 × 3
## orig.col.name Species foo
## <chr> <fct> <dbl>
## 1 Sepal.Length setosa 5.01
## 2 Sepal.Length versicolor 5.94
## 3 Sepal.Length virginica 6.59
## 4 Sepal.Width setosa 3.43
## 5 Sepal.Width versicolor 2.77
## 6 Sepal.Width virginica 2.97
## 7 Petal.Length setosa 1.46
## 8 Petal.Length versicolor 4.26
## 9 Petal.Length virginica 5.55
## 10 Petal.Width setosa 0.246
## 11 Petal.Width versicolor 1.33
## 12 Petal.Width virginica 2.03
collapse::pivot(N.tall.df, how="w", ids=c("orig.col.name","Species"), values="value", names="name", FUN=mean)
## orig.col.name Species foo
## 1 Sepal.Length setosa 5.006
## 2 Sepal.Length versicolor 5.936
## 3 Sepal.Length virginica 6.588
## 4 Sepal.Width setosa 3.428
## 5 Sepal.Width versicolor 2.770
## 6 Sepal.Width virginica 2.974
## 7 Petal.Length setosa 1.462
## 8 Petal.Length versicolor 4.260
## 9 Petal.Length virginica 5.552
## 10 Petal.Width setosa 0.246
## 11 Petal.Width versicolor 1.326
## 12 Petal.Width virginica 2.026
DBI::dbGetQuery(con, 'PIVOT iris_tall USING mean(value) GROUP BY "orig.col.name", "Species"')
## orig.col.name Species mean("value")
## 1 Sepal.Length versicolor 5.936000
## 2 Sepal.Width setosa 3.428008
## 3 Sepal.Width virginica 2.974000
## 4 Petal.Length versicolor 4.260000
## 5 Petal.Width setosa 0.245998
## 6 Petal.Width virginica 2.026000
## 7 Sepal.Length setosa 5.006010
## 8 Sepal.Length virginica 6.588000
## 9 Sepal.Width versicolor 2.770000
## 10 Petal.Length setosa 1.462000
## 11 Petal.Length virginica 5.552000
## 12 Petal.Width versicolor 1.326000
Note in the code above that to get the tidyr and collapse methods
to work, a “name” column must be created, whereas in data.table it
not necessary (you just specify . in right side of formula to
indicate that only one column should be output). The table below summarizes the differences in syntax between the different R functions:
| function | ID | Name | Value | Aggregation |
|---|---|---|---|---|
data.table::dcast |
LHS of formula | RHS of formula | value.var |
fun.aggregate |
stats::aggregate |
by |
all column names | all input values | FUN |
tidyr::pivot_wider |
id_cols |
names_from |
values_from |
values_fn |
collapse::pivot |
ids |
names |
values |
FUN |
polars $pivot |
index |
on |
values |
- |
duckdb PIVOT |
GROUP BY |
ON |
USING |
USING |
UPDATE 27 Sept 2024: added duckdb based on docs overview, pivot, unpivot. Note that duckdb supports multiple value columns, and multiple aggregation functions, but you need to specify each combination of column/aggregation in a separate entry of USING, so this is less convenient than data.table::dcast, which computes each aggregation function for each value column.
Below we compare the performance of these different functions.
m.res <- atime::atime(
N=2^seq(1,50),
setup={
(row.id.vec <- 1+(seq(0,N-1) %% nrow(iris)))
N.df <- iris[row.id.vec,]
N.tall.df <- reshape_taller(N.df,1:4)
N.tall.df$name <- "foo"
N.tall.dt <- data.table(N.tall.df)
},
seconds.limit=0.1,
##"duckdb\ndbWriteTable"=duckdb::dbWriteTable(con, "iris_tall", N.tall.df, overwrite=TRUE),
##"duckdb\nPIVOT"=DBI::dbGetQuery(con, 'PIVOT iris_tall USING mean(value) GROUP BY "orig.col.name", "Species"'),
"stats\naggregate"=stats::aggregate(N.tall.df[,"value",drop=FALSE], by=with(N.tall.df, list(orig.col.name=orig.col.name,Species=Species)), FUN=mean),
"tidyr\npivot_wider"=tidyr::pivot_wider(N.tall.df, names_from=name, values_from=value, id_cols=c(orig.col.name,Species), values_fn=mean),
"collapse\npivot"=collapse::pivot(N.tall.df, how="w", ids=c("orig.col.name","Species"), values="value", names="name", FUN=mean),
"data.table\ndcast"=dcast(N.tall.dt, orig.col.name + Species ~ ., mean))
m.refs <- atime::references_best(m.res)
m.pred <- predict(m.refs)
plot(m.pred)+coord_cartesian(xlim=c(NA,1e8))
## Warning in ggplot2::scale_x_log10("N", breaks = meas[, 10^seq(ceiling(min(log10(N))), : log-10 transformation
## introduced infinite values.
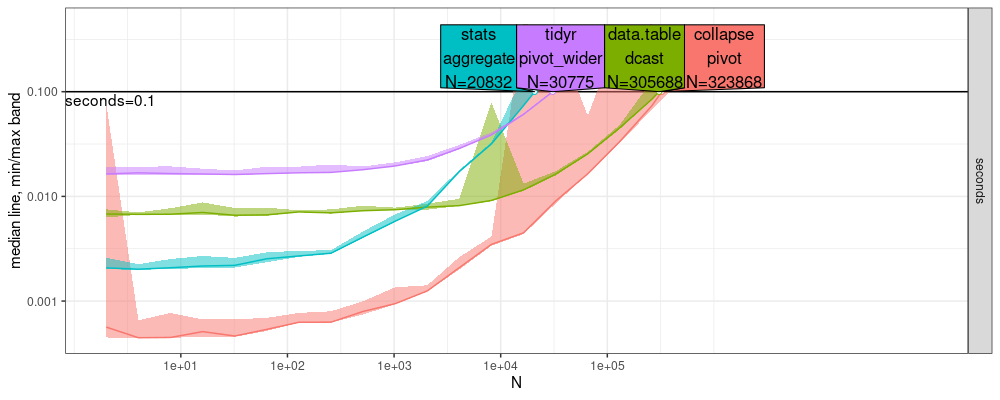
The result above shows that data.table is a bit faster than
collapse. Upon close inspection of the log-log plot above, we see
that data.table has a smaller slope than collapse, which means
that `data.table has an asymptotically more efficient complexity
class. To estimate the asymptotic complexity class, we can use the
plot below.
m.refs$plot.references <- m.refs$ref[fun.name %in% c("N","N log N")]
plot(m.refs)
## Warning in ggplot2::scale_y_log10(""): log-10 transformation introduced infinite values.
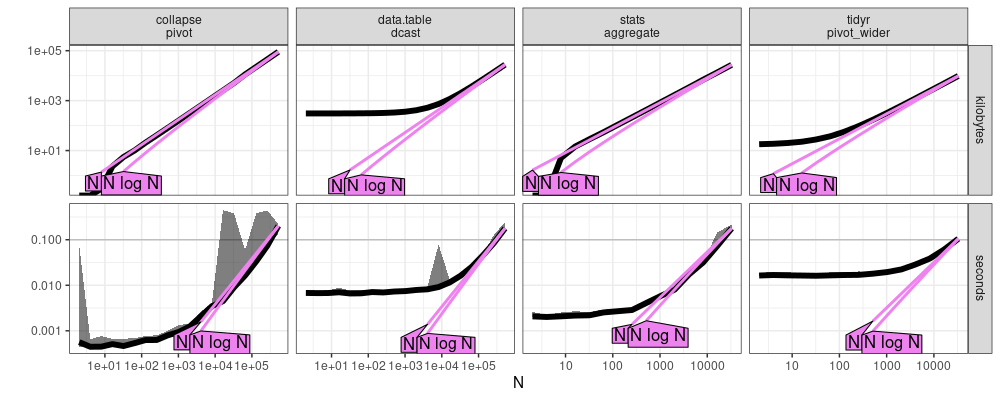
The plot above indicates that all methods use asymptotically linear
memory, O(N), in the number of input rows N. It also suggests
that collapse::pivot and stats::aggregate may be asymptotically log-linear time,
O(N log N), whereas data.table::dcast and tidyr::pivot_wider are clearly linear time,
O(N).
Multiple aggregation functions
It is often useful to provide a list of aggregation functions, instead of just one. For example, in visualization results of machine learning predictions, we would like to use
meanto compute the mean prediction error over several train/test splits,sdto get the standard deviation,lengthto double-check that the number of items to summarize is the same as the number of cross-validation folds.
In data.table we can provide a list of functions as in the code below,
dcast(N.tall.dt, orig.col.name + Species ~ ., list(mean, sd, length))
## Key: <orig.col.name, Species>
## orig.col.name Species value_mean value_sd value_length
## <char> <fctr> <num> <num> <int>
## 1: Petal.Length setosa 1.462 0.1727847 100
## 2: Petal.Length versicolor 4.260 0.4675317 100
## 3: Petal.Length virginica 5.552 0.5518947 50
## 4: Petal.Width setosa 0.246 0.1048520 100
## 5: Petal.Width versicolor 1.326 0.1967514 100
## 6: Petal.Width virginica 2.026 0.2746501 50
## 7: Sepal.Length setosa 5.006 0.3507049 100
## 8: Sepal.Length versicolor 5.936 0.5135576 100
## 9: Sepal.Length virginica 6.588 0.6358796 50
## 10: Sepal.Width setosa 3.428 0.3771450 100
## 11: Sepal.Width versicolor 2.770 0.3122095 100
## 12: Sepal.Width virginica 2.974 0.3224966 50
However this feature is not supported in other packages.
?stats::aggregatesays “FUN: a function to compute the summary statistics which can be applied to all data subsets.”?collapse::pivotsays “FUN: function to aggregate values. At present, only a single function is allowed.”?tidyr::pivot_widersaysvalues_fnargument “can be a named list if you want to apply different aggregations to differentvalues_fromcolumns.”
Conclusion
We have shown how to use atime to check if there are any performance
differences between data reshaping functions in R. We observed large
differences between functions when doing a reshape in the wider
direction, with no aggregation (collapse is about 50x faster than
data.table for this case, which involves just copying values to a
new table). For reshape in the tall/long direction, and for reshape
wider with aggregation, we observed small constant factor differences
between methods (collapse was still fastest but by only 2x).
In terms of functionality, we observed a few differences. Wide-to-tall
reshape defined by a regex is supported by
tidyr::pivot_longer(names_pattern=regex) and
data.table::measure(pattern=regex), whereas stats::reshape only
supports multiple outputs via the sep argument (separator, not
regex), and collapse::pivot does not support multiple outputs at
all. Also, we observed that data.table::dcast is the only function
that supports multiple aggregation functions, which can be useful for
simultaneously computing a variety of summary statistics, such as
mean, sd, and length. The table below summarizes these
differences in functionality.
| reshape long | reshape long | reshape wide | reshape wide | reshape wide | |
|---|---|---|---|---|---|
| package | multiple outputs | using regex | multiple agg. | single agg. | no agg. |
data.table |
yes | yes | yes | O(N) | O(N) |
tidyr |
yes | yes | no | O(N) | O(N log N)? |
stats |
yes | no | no | O(N log N)? | O(N) |
collapse |
no | no | no | O(N log N)? | O(N log N)? |
polars |
no | no | no | no support | O(N log N)? |
duckdb |
no | no | yes | O(N)? | O(N)? |
UPDATE 26 sept 2024 added polars row based on docs for pivot and unpivot.
For future work,
data.table/tidyrmay consider trying to speed up the constant factors in the reshape wide code without aggregation.collapsereshape wide with aggregation seems asymptotically log-linear, and so may consider investigating the possibility of a speedup to asymptotically linear.tidyr/collapse/polarsmay consider implementing the advanced reshaping features thatdata.tablecurrently supports (multiple outputs, regex).
Session info
sessionInfo()
## R version 4.4.1 (2024-06-14)
## Platform: x86_64-pc-linux-gnu
## Running under: Ubuntu 22.04.5 LTS
##
## Matrix products: default
## BLAS: /usr/lib/x86_64-linux-gnu/blas/libblas.so.3.10.0
## LAPACK: /usr/lib/x86_64-linux-gnu/lapack/liblapack.so.3.10.0
##
## locale:
## [1] LC_CTYPE=fr_FR.UTF-8 LC_NUMERIC=C LC_TIME=fr_FR.UTF-8 LC_COLLATE=fr_FR.UTF-8
## [5] LC_MONETARY=fr_FR.UTF-8 LC_MESSAGES=fr_FR.UTF-8 LC_PAPER=fr_FR.UTF-8 LC_NAME=C
## [9] LC_ADDRESS=C LC_TELEPHONE=C LC_MEASUREMENT=fr_FR.UTF-8 LC_IDENTIFICATION=C
##
## time zone: America/New_York
## tzcode source: system (glibc)
##
## attached base packages:
## [1] stats graphics utils datasets grDevices methods base
##
## other attached packages:
## [1] data.table_1.16.99 ggplot2_3.5.1
##
## loaded via a namespace (and not attached):
## [1] gtable_0.3.4 crayon_1.5.2 dplyr_1.1.4 compiler_4.4.1 highr_0.11
## [6] tidyselect_1.2.1 Rcpp_1.0.12 collapse_2.0.15 parallel_4.4.1 tidyr_1.3.1
## [11] directlabels_2024.1.21 scales_1.3.0 fastmap_1.1.1 lattice_0.22-6 R6_2.5.1
## [16] labeling_0.4.3 generics_0.1.3 knitr_1.47 tibble_3.2.1 polars_0.19.1
## [21] munsell_0.5.0 atime_2024.9.27 DBI_1.2.1 pillar_1.9.0 rlang_1.1.3
## [26] utf8_1.2.4 xfun_0.45 quadprog_1.5-8 cli_3.6.2 withr_3.0.0
## [31] magrittr_2.0.3 digest_0.6.34 grid_4.4.1 nc_2024.9.19 lifecycle_1.0.4
## [36] vctrs_0.6.5 bench_1.1.3 evaluate_0.23 glue_1.7.0 farver_2.1.1
## [41] duckdb_1.1.0 profmem_0.6.0 fansi_1.0.6 colorspace_2.1-0 rmarkdown_2.25.1
## [46] purrr_1.0.2 tools_4.4.1 pkgconfig_2.0.3 htmltools_0.5.7