Benchmarking data.table with polars, duckdb, and pandas
The goal of this post is to demonstrate that the performance of
data.table CSV readers/writers is competitive with similar libraries
available in Python (pandas, duckdb, polars). Compare with the
previous post,
which compared R functions (not python).
What is the overhead of communication with python from R?
We will be recording the timings of python code from R, via the reticulate package. To use that, we must tell reticulate where to find a python (conda environment) which has pandas installed:
reticulate::use_condaenv("~/miniconda3/envs/2023-08-deep-learning/bin/python3.11")
If you want to reproduce the results in this blog on your own computer, make sure to change the line above to a valid conda environment.
Below, we run a test to determine what is the overhead involved in communicating with python from R via reticulate.
atime.overhead <- atime::atime(
N = as.integer(10^seq(0, 3, by = 0.5)),
overhead = {
reticulate::py_run_string("x = 5")
}
)
atime.overhead$unit.col.vec <- c(seconds="median")
plot(atime.overhead)
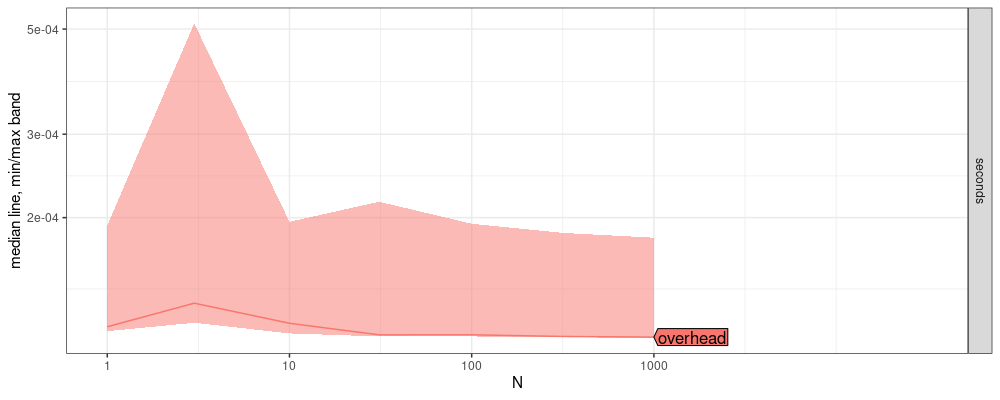
(overhead.seconds <- median(atime.overhead$measurements$median))
## [1] 0.0001131966
The plot above shows that the overhead of communication between R and python is about 0.1131966 milliseconds.
write csv, vary columns, char
This section compares the performance of CSV files with a variable number of character/string columns..
library(data.table)
reticulate::py_run_string("import pandas as pd")
N_rows <- 100
(N_col_seq <- unique(as.integer(10^seq(0, 6, by = 0.2))))
## [1] 1 2 3 6 10 15 25 39 63 100 158 251 398 630
## [15] 1000 1584 2511 3981 6309 10000 15848 25118 39810 63095 100000 158489 251188 398107
## [29] 630957 1000000
main <- reticulate::py
con <- duckdb::dbConnect(duckdb::duckdb(), dbdir = ":memory:")
atime.write.chr.cols <- atime::atime(
N = N_col_seq,
setup = {
input.chr <- matrix(c("foo", "x", "bar", NA), N_rows, N)
input.df <- data.frame(input.chr)
polars.df <- polars::as_polars_df(input.df)
main$input_df_pd <- input.df
input.dt <- data.table(input.df)
duckdb::dbWriteTable(con, "duck_table", input.df, overwrite=TRUE)
out.csv <- sprintf("%s/data_N=%d.csv", tempdir(), N)
},
seconds.limit = 0.1,
"data.table\nfwrite" = {
data.table::fwrite(input.dt, out.csv, showProgress = FALSE)
},
"pandas\nto_csv" = {
reticulate::py_run_string("input_df_pd.to_csv('data.csv', index=False)")
},
"polars\nto_csv" = {
polars.df$write_csv('polars.csv')
},
"duckdb\nCOPY" = { # https://duckdb.org/docs/guides/file_formats/csv_export.html
DBI::dbSendQuery(con, "COPY duck_table TO 'duck.csv' (HEADER, DELIMITER ',')")
}
)
atime.write.chr.cols$unit.col.vec <- c(seconds="median")
library(ggplot2)
plot(atime.write.chr.cols)+
geom_hline(aes(
yintercept=overhead.seconds),
color="grey50",
data=data.frame(overhead.seconds))
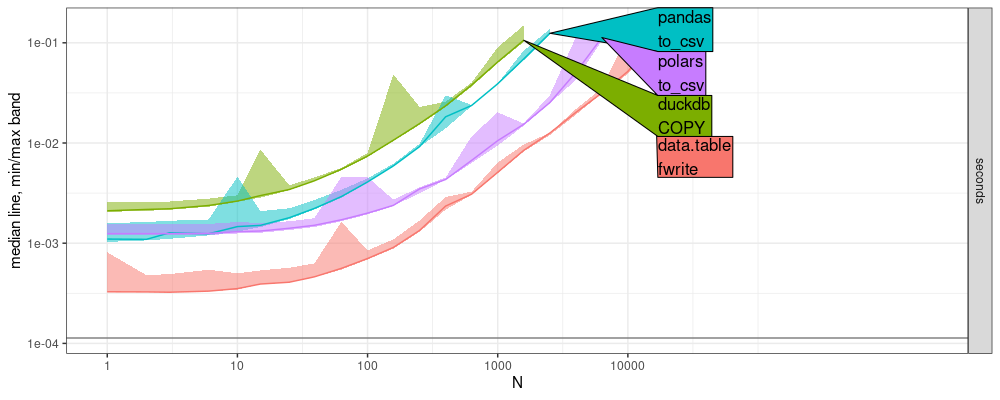
The plot above shows a colored curve for each CSV writing function. The grey horizontal line shows the overhead of communication with python from R. We see that the overhead (grey line) is insignificant, compared with the CSV writing timings (colored curves). The code below shows the throughput of each function at the specified time limit.
refs.write.chr.cols <- atime::references_best(atime.write.chr.cols)
pred.write.chr.cols <- predict(refs.write.chr.cols)
plot(pred.write.chr.cols)+
ggtitle("Write strings to CSV, 100 x N")+
scale_x_log10("N = number of columns to write")+
scale_y_log10("Computation time (seconds)\nmedian line, min/max band\nover 10 timings",limits=c(1e-4,0.5))+
facet_null()
## Scale for x is already present.
## Adding another scale for x, which will replace the existing scale.
## Scale for y is already present.
## Adding another scale for y, which will replace the existing scale.
## Warning in scale_x_log10("N = number of columns to write"): log-10 transformation introduced infinite values.
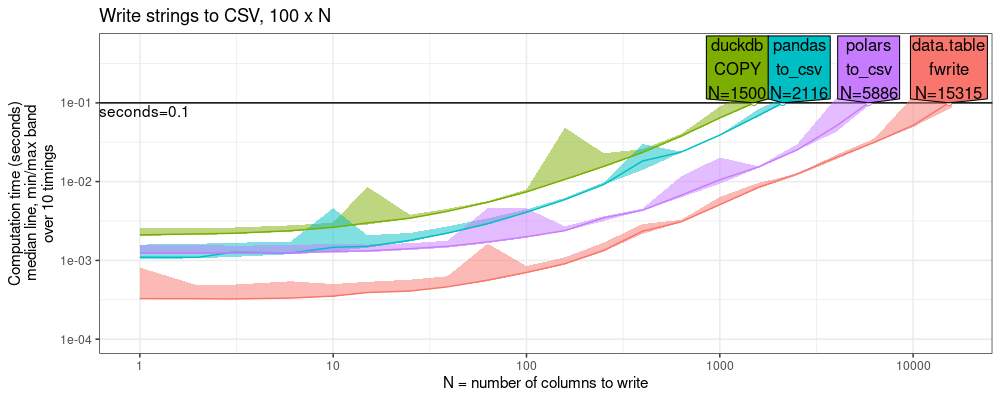
The plot above shows that data.table is competitive with the other
libraries: about 2x faster than polars, and 10x faster than pandas/duckdb.
read CSV, vary columns, char
In this section we compare performance of CSV readers.
atime.read.chr.cols <- atime::atime(
N = N_col_seq,
setup = {
in.csv <- sprintf("%s/data_N=%d.csv", tempdir(), N)
pandas_cmd <- sprintf("pd.read_csv('%s')", in.csv)
## https://duckdb.org/docs/data/csv/overview.html
duck_cmd <- sprintf("SELECT * FROM read_csv('%s', delim=',', header=true)", in.csv)
},
seconds.limit = 0.1,
"data.table\nfread" = {
data.table::fread(in.csv, showProgress = FALSE)
},
"pandas\nread_csv" = {
reticulate::py_run_string(pandas_cmd)
},
"duckdb\nread_csv" = {
DBI::dbSendQuery(con, duck_cmd)
},
"polars\nread_csv" = {
polars::pl$read_csv(in.csv)
}
)
refs.read.chr.cols <- atime::references_best(atime.read.chr.cols)
pred.read.chr.cols <- predict(refs.read.chr.cols)
plot(pred.read.chr.cols)+
ggtitle("Read strings from CSV, 100 x N")+
scale_x_log10("N = number of columns to read")+
scale_y_log10("Computation time (seconds)\nmedian line, min/max band\nover 10 timings",limits=c(1e-4,0.5))+
facet_null()
## Scale for x is already present.
## Adding another scale for x, which will replace the existing scale.
## Scale for y is already present.
## Adding another scale for y, which will replace the existing scale.
## Warning in scale_x_log10("N = number of columns to read"): log-10 transformation introduced infinite values.
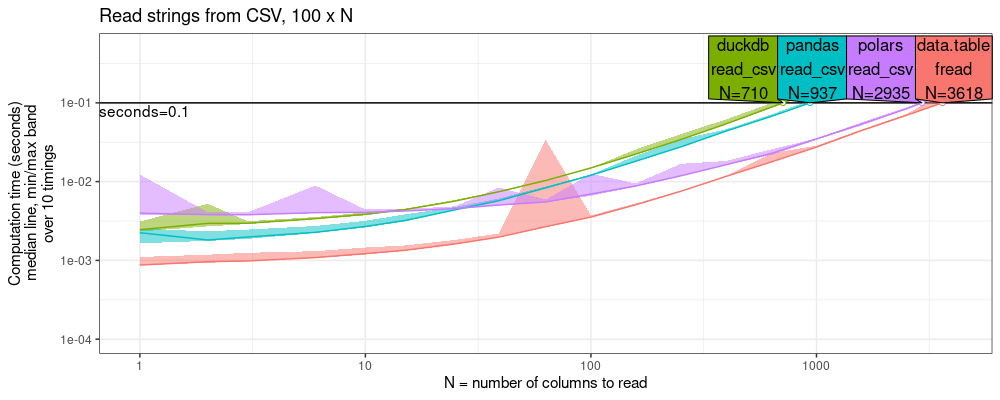
The plot above shows that data.table has the fastest CSV reader.
The plot above shows the throughput at the time limit, which reveals
that data.table is faster than polars by a small factor (less than
2x), but faster than duckdb/pandas by a larger amount (3-5x).
variants of these benchmarks
There are several variables we can play with in these benchmarks.
- variable number of rows or columns (we did columns above)
- numeric or character/string data (we did character/string above)
- read or write (we did both above)
Below we consider two variants of the benchmark.
write csv, variable rows
Here we consider writing a variable number of rows (instead of cols above) for character/string data. (numeric data and reading are exercises for the reader)
atime.write.chr.rows <- atime::atime(
setup = {
input.chr <- matrix(c("foo", "x", "bar", NA), N, 10)
input.df <- data.frame(input.chr)
polars.df <- polars::as_polars_df(input.df)
main$input_df_pd <- input.df
input.dt <- data.table(input.df)
duckdb::dbWriteTable(con, "duck_table", input.df, overwrite=TRUE)
out.csv <- sprintf("%s/data_N=%d.csv", tempdir(), N)
},
seconds.limit = 0.01,
"data.table\nfwrite" = {
data.table::fwrite(input.dt, out.csv, showProgress = FALSE)
},
"pandas\nto_csv" = {
reticulate::py_run_string("input_df_pd.to_csv('data.csv', index=False)")
},
"polars\nto_csv" = {
polars.df$write_csv('polars.csv')
},
"duckdb\nCOPY" = { # https://duckdb.org/docs/guides/file_formats/csv_export.html
DBI::dbSendQuery(con, "COPY duck_table TO 'duck.csv' (HEADER, DELIMITER ',')")
}
)
refs.write.chr.rows <- atime::references_best(atime.write.chr.rows)
pred.write.chr.rows <- predict(refs.write.chr.rows)
plot(pred.write.chr.rows)+
ggtitle("Write strings to CSV, N x 100")+
scale_y_log10("Computation time (seconds)\nmedian line, min/max band\nover 10 timings")+
facet_null()+
scale_x_log10("N = number of rows to write")
## Scale for y is already present.
## Adding another scale for y, which will replace the existing scale.
## Scale for x is already present.
## Adding another scale for x, which will replace the existing scale.
## Warning in scale_x_log10("N = number of rows to write"): log-10 transformation introduced infinite values.
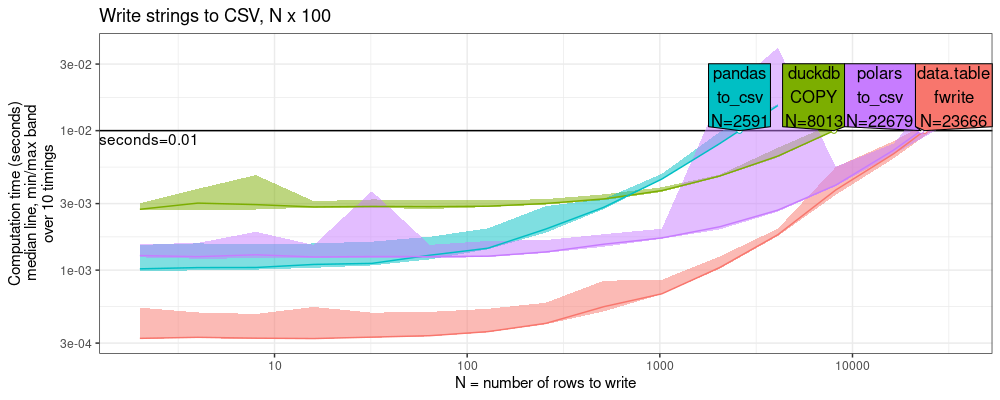
The plot above again shows that data.table is the fastest CSV writer.
It is about the same as polars (within 2x),
about 3x faster than duckdb,
and 10x faster than pandas.
write csv, numeric cols
Here we consider writing a variable number of cols for numeric data (instead of character/string data above). Variable number of rows, and reading instead of writing, are exercises for the reader.
atime.write.num.cols <- atime::atime(
N = N_col_seq,
setup = {
set.seed(1)
input.num <- matrix(rnorm(N_rows * N), N_rows, N)
input.num[sample(N_rows*N, N)] <- NA
input.df <- data.frame(input.num)
polars.df <- polars::as_polars_df(input.df)
main$input_df_pd <- input.df
input.dt <- data.table(input.df)
duckdb::dbWriteTable(con, "duck_table", input.df, overwrite=TRUE)
out.csv <- sprintf("%s/data_N=%d.csv", tempdir(), N)
},
seconds.limit = 0.1,
"data.table\nfwrite" = {
data.table::fwrite(input.dt, out.csv, showProgress = FALSE)
},
"pandas\nto_csv" = {
reticulate::py_run_string("input_df_pd.to_csv('data.csv', index=False)")
},
"polars\nto_csv" = {
polars.df$write_csv('polars.csv')
},
"duckdb\nCOPY" = { # https://duckdb.org/docs/guides/file_formats/csv_export.html
DBI::dbSendQuery(con, "COPY duck_table TO 'duck.csv' (HEADER, DELIMITER ',')")
}
)
refs.write.num.cols <- atime::references_best(atime.write.num.cols)
pred.write.num.cols <- predict(refs.write.num.cols)
plot(pred.write.num.cols)+
ggtitle("Write real numbers to CSV, 100 x N")+
scale_y_log10("Computation time (seconds)\nmedian line, min/max band\nover 10 timings",limits=c(1e-4,0.5))+
facet_null()+
scale_x_log10("N = number of columns to write")
## Scale for y is already present.
## Adding another scale for y, which will replace the existing scale.
## Scale for x is already present.
## Adding another scale for x, which will replace the existing scale.
## Warning in scale_x_log10("N = number of columns to write"): log-10 transformation introduced infinite values.
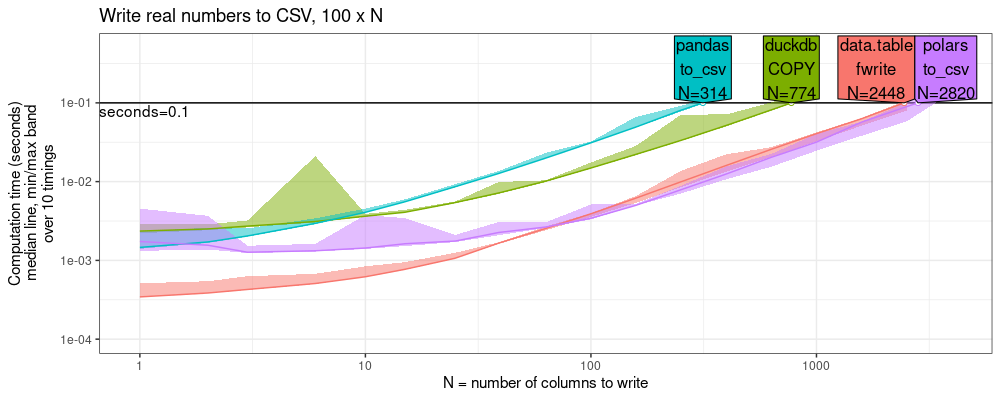
The code above shows the throughput of each function at the specified time limit.
The plot above shows that data.table is competitive with the other
libraries: almost as fast as polars, and several times faster than
pandas/duckdb.
This indicates that data.table is especially efficient for
character/string data (rather than numeric data).
Summarize by group
In this section, we compare the time required for computing a summarization. In machine learning we often run gradient descent for several epochs, and we want to average the loss over cross-validation folds. The experiment below assumes we have 10 cross-validation folds, and N epochs.
n.folds <- 10
ml.atime <- atime::atime(
setup={
set.seed(1)
loss.dt <- data.table(
name="loss",
fold=rep(1:n.folds, each=N),
loss=rnorm(N*n.folds),
set=rep(c("subtrain","validation"),each=N/2),
epoch=seq(1,N/2),
key=c("set","epoch","fold"))
loss.polars <- polars::as_polars_df(loss.dt)
main$loss_df_pd <- loss.dt
duckdb::dbWriteTable(con, "loss_table", loss.dt, overwrite=TRUE)
},
seconds.limit=0.1,
data.table={
loss.dt[, .(
loss_length=.N,
loss_mean=mean(loss),
loss_sd=sd(loss)
), by=.(set, epoch)]
},
polars={
loss.polars$group_by(c("set","epoch"))$agg(
loss_len=polars::pl$col("loss")$len(),
loss_mean=polars::pl$col("loss")$mean(),
loss_std=polars::pl$col("loss")$std())
},
pandas={
reticulate::py_run_string("loss_df_pd.groupby(['set','epoch']).loss.agg(['size','mean','std'])")
},
duckdb={
DBI::dbSendQuery(con, "
SELECT set, epoch,
count(*) as loss_count,
mean(loss) as loss_mean,
stddev(loss) as loss_stddev
FROM loss_table
GROUP BY set, epoch")
})
ml.refs <- atime::references_best(ml.atime)
ml.pred <- predict(ml.refs)
plot(ml.pred)+
ggtitle(sprintf("Mean,SD,Length over %d real numbers, N times", n.folds))+
scale_x_log10("N = number of Mean,SD,Length to compute")+
scale_y_log10("Computation time (seconds)\nmedian line, min/max band\nover 10 timings")+
facet_null()
## Scale for x is already present.
## Adding another scale for x, which will replace the existing scale.
## Scale for y is already present.
## Adding another scale for y, which will replace the existing scale.
## Warning in scale_x_log10("N = number of Mean,SD,Length to compute"): log-10 transformation introduced infinite values.
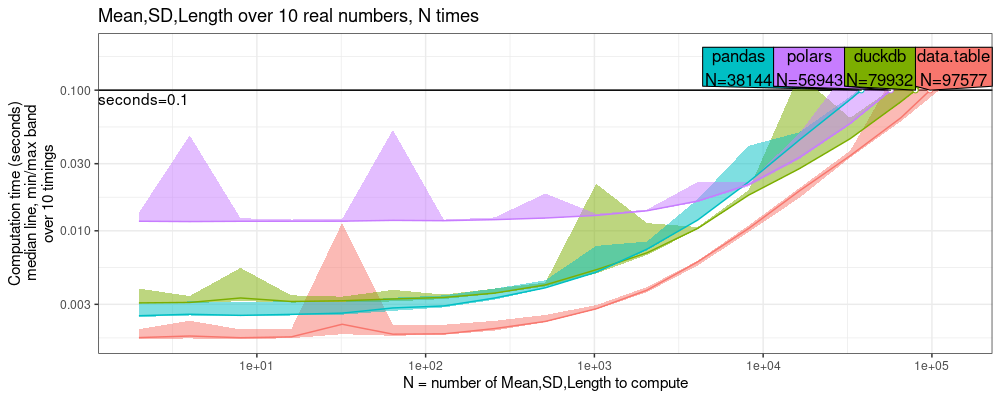
The result above shows that data.table is 2-3x faster than the other software.
Conclusions
We have shown that data.table can be faster
than similar libraries which can be used from python or R.
Session info
sessionInfo()
## R Under development (unstable) (2024-10-01 r87205)
## Platform: x86_64-pc-linux-gnu
## Running under: Ubuntu 22.04.5 LTS
##
## Matrix products: default
## BLAS: /usr/lib/x86_64-linux-gnu/blas/libblas.so.3.10.0
## LAPACK: /home/tdhock/miniconda3/envs/2023-08-deep-learning/lib/libmkl_rt.so.2; LAPACK version 3.10.1
##
## locale:
## [1] LC_CTYPE=fr_FR.UTF-8 LC_NUMERIC=C LC_TIME=fr_FR.UTF-8 LC_COLLATE=fr_FR.UTF-8
## [5] LC_MONETARY=fr_FR.UTF-8 LC_MESSAGES=fr_FR.UTF-8 LC_PAPER=fr_FR.UTF-8 LC_NAME=C
## [9] LC_ADDRESS=C LC_TELEPHONE=C LC_MEASUREMENT=fr_FR.UTF-8 LC_IDENTIFICATION=C
##
## time zone: America/New_York
## tzcode source: system (glibc)
##
## attached base packages:
## [1] stats graphics utils datasets grDevices methods base
##
## other attached packages:
## [1] ggplot2_3.5.1 data.table_1.16.2
##
## loaded via a namespace (and not attached):
## [1] Matrix_1.7-0 gtable_0.3.4 jsonlite_1.8.8 dplyr_1.1.4 compiler_4.5.0
## [6] highr_0.11 tidyselect_1.2.1 Rcpp_1.0.12 scales_1.3.0 png_0.1-8
## [11] directlabels_2024.1.21 reticulate_1.34.0.9000 lattice_0.22-6 R6_2.5.1 labeling_0.4.3
## [16] generics_0.1.3 knitr_1.47 tibble_3.2.1 polars_0.19.1 munsell_0.5.0
## [21] atime_2024.12.3 DBI_1.2.1 pillar_1.9.0 rlang_1.1.3 utf8_1.2.4
## [26] xfun_0.45 quadprog_1.5-8 cli_3.6.2 withr_3.0.0 magrittr_2.0.3
## [31] grid_4.5.0 lifecycle_1.0.4 vctrs_0.6.5 bench_1.1.3 evaluate_0.23
## [36] glue_1.7.0 farver_2.1.1 duckdb_1.1.0 profmem_0.6.0 fansi_1.0.6
## [41] colorspace_2.1-0 tools_4.5.0 pkgconfig_2.0.3
reticulate::py_run_string("print(pd.__version__)")
## 2.1.1