Markdown links to bibtex
The purpose of this page is to convert markdown links to bibtex using regular expressions.
Problem
I am using markdown (actually quarto) to write two online “books”
- Animated interactive data visualization using animint2 in R
- Visualisation interactive de données dans R avec animint2
Including hyperlinks in markdown is via the syntax
[text that will appear](hyperlink)
This works fine for HTML output, where we can click the link. But in PDF output we get blue text, and the link is lost when the document is printed. An example is shown below.
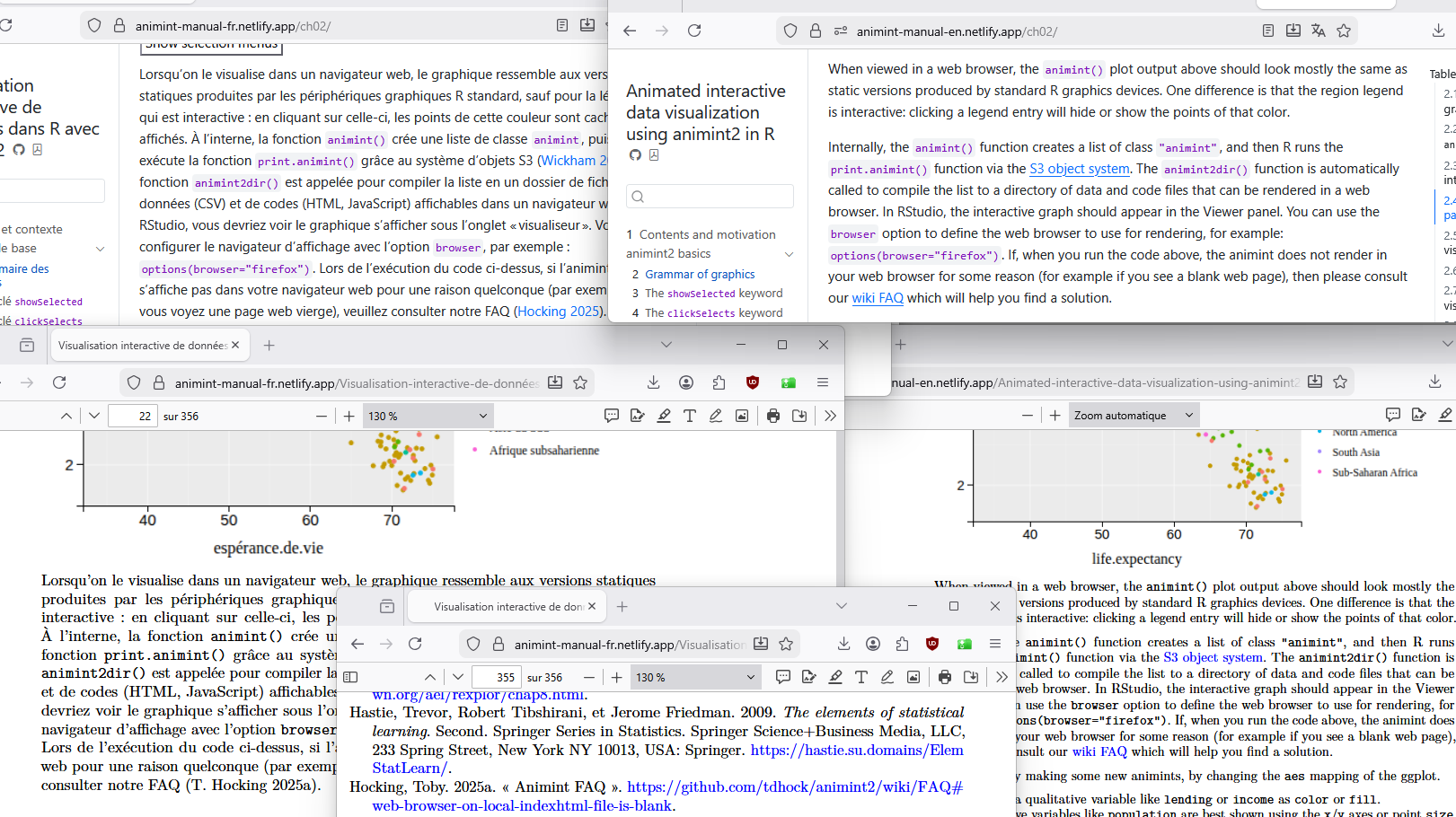
- left we see the French version with bibtex references.
- right we see the English version with markdown hyperlinks.
- top we see the web page output.
- bottom we see the PDF output (and https URL in references section of French version).
I already wrote a CI script test-references.R that errors if it finds markdown links.
chapters <- "~/R/animint-manual-en/chapters"
qmd.files <- c(
Sys.glob(file.path(chapters,"*qmd")),
Sys.glob(file.path(chapters,"*/index.qmd")))
library(data.table)
## data.table 1.18.2.1 utilise 1 threads (voir ?getDTthreads). Dernières actualités : r-datatable.com
## **********
## Exécution de data.table en anglais ; l'aide du package n'est disponible qu'en anglais. Lorsque vous recherchez de l'aide en ligne, veillez à vérifier également le message d'erreur en anglais. Pour ce faire, consultez les fichiers po/R-<locale>.po et po/<locale>.po dans le source du package, où les messages d'erreur en langue native et ceux en anglais figurent côte à côte. Vous pouvez aussi essayer d'appeler Sys.setLanguage('en') avant de reproduire le message d'erreur.
## **********
##
## Attachement du package : 'data.table'
##
## L'objet suivant est masqué depuis 'package:base':
##
## %notin%
get_violations <- function(qmd){
nc::capture_all_str(
qmd,
"\\]\\(",
url="http.*?",
"\\)")$url
}
violations <- list()
for(qmd in qmd.files){
bad <- get_violations(qmd)
if(length(bad)){
violations[[qmd]] <- bad
}
}
print(violations)
## $`/home/local/USHERBROOKE/hoct2726/R/animint-manual-en/chapters/contribute.qmd`
## [1] "https://github.com/animint/animint-manual-en/issues/new"
##
## $`/home/local/USHERBROOKE/hoct2726/R/animint-manual-en/chapters/index.qmd`
## [1] "https://en.wikipedia.org/wiki/Lady_tasting_tea"
## [2] "https://en.wikipedia.org/wiki/Keeling_Curve"
##
## $`/home/local/USHERBROOKE/hoct2726/R/animint-manual-en/chapters/ch02/index.qmd`
## [1] "https://en.wiktionary.org/wiki/sketch"
## [2] "http://adv-r.had.co.nz/OO-essentials.html#s3"
## [3] "https://github.com/tdhock/animint2/wiki/FAQ#web-browser-on-local-indexhtml-file-is-blank"
##
## $`/home/local/USHERBROOKE/hoct2726/R/animint-manual-en/chapters/ch03/index.qmd`
## [1] "https://yihui.name/animation/examples/"
##
## $`/home/local/USHERBROOKE/hoct2726/R/animint-manual-en/chapters/ch05/index.qmd`
## [1] "http://rmarkdown.rstudio.com/"
## [2] "https://app.netlify.com/drop"
## [3] "https://pages.github.com/"
## [4] "https://github.com/tdhock/animint2/wiki/FAQ#web-browser-on-local-indexhtml-file-is-blank"
## [5] "https://app.netlify.com/drop"
## [6] "https://docs.netlify.com/deploy/deploy-overview/"
## [7] "https://pages.github.com/"
## [8] "https://github.com/join"
## [9] "https://github.com/tdhock/cs499-spring2020"
## [10] "https://github.com/tdhock/cs499-spring2020/blob/master/2020-02-03-capacity/figure-quadratic-interactive.R"
## [11] "https://vimeo.com"
## [12] "https://github.com/tdhock/cs499-spring2020/blob/master/2020-02-03-capacity/figure-several-interactive.R"
## [13] "https://github.com/tdhock/cs499-spring2020/blob/master/2020-02-03-capacity/figure-quadratic-interactive.R"
## [14] "https://github.com/tdhock/2020-02-03-capacity-polynomial-degree/blob/gh-pages/README.md"
## [15] "https://github.com/animint/gallery/blob/gh-pages/repos.txt"
##
## $`/home/local/USHERBROOKE/hoct2726/R/animint-manual-en/chapters/ch06/index.qmd`
## [1] "http://www.w3schools.com/html/html5_svg.asp"
## [2] "http://www.w3schools.com/tags/att_global_id.asp"
## [3] "https://github.com/tdhock/animint/wiki/Testing"
## [4] "https://tdhock.github.io/2025-01-WorldBank-facets-map/"
## [5] "https://tdhock.github.io/2025-01-WorldBank-facets-map/"
## [6] "https://tdhock.github.io/figure-binseg-cv-most-frequently-selected-fr/"
## [7] "http://selectize.github.io/selectize.js/"
## [8] "https://github.com/tdhock/animint2/blob/master/tests/testthat/test-renderer2-PredictedPeaks.R"
##
## $`/home/local/USHERBROOKE/hoct2726/R/animint-manual-en/chapters/ch07/index.qmd`
## [1] "https://github.com/tdhock/animint2/compare"
## [2] "https://github.com/tdhock/animint2/blob/master/tests/testthat/test-renderer4-update-axes-multiple-ss.R"
## [3] "https://github.com/tdhock/animint/issues/148"
## [4] "https://github.com/tdhock/animint/issues/149"
## [5] "https://github.com/animint/animint2/issues/230"
## [6] "http://docs.ggplot2.org/current/theme.html"
##
## $`/home/local/USHERBROOKE/hoct2726/R/animint-manual-en/chapters/ch08/index.qmd`
## [1] "https://tdhock.github.io/2025-01-WorldBank-facets-map/"
##
## $`/home/local/USHERBROOKE/hoct2726/R/animint-manual-en/chapters/ch10/index.qmd`
## [1] "http://statweb.stanford.edu/~tibs/ElemStatLearn/"
##
## $`/home/local/USHERBROOKE/hoct2726/R/animint-manual-en/chapters/ch11/index.qmd`
## [1] "https://en.wikipedia.org/wiki/Lasso_%28statistics%29"
##
## $`/home/local/USHERBROOKE/hoct2726/R/animint-manual-en/chapters/ch12/index.qmd`
## [1] "https://en.wikipedia.org/wiki/Support_vector_machine"
##
## $`/home/local/USHERBROOKE/hoct2726/R/animint-manual-en/chapters/ch13/index.qmd`
## [1] "https://en.wikipedia.org/wiki/Poisson_regression"
## [2] "https://en.wikipedia.org/wiki/Binomial_regression"
##
## $`/home/local/USHERBROOKE/hoct2726/R/animint-manual-en/chapters/ch14/index.qmd`
## [1] "https://github.com/tdhock/PeakSegJoint"
## [2] "https://tdhock.github.io/2023-12-04-degree-neighbors/"
##
## $`/home/local/USHERBROOKE/hoct2726/R/animint-manual-en/chapters/ch15/index.qmd`
## [1] "https://www.wolframalpha.com/input/?i=a*x+%2Bb*log%28x%29%2B+c%3D0"
## [2] "https://en.wikipedia.org/wiki/Newton%27s_method"
## [3] "https://en.wikipedia.org/wiki/Binomial_regression"
##
## $`/home/local/USHERBROOKE/hoct2726/R/animint-manual-en/chapters/ch16/index.qmd`
## [1] "https://github.com/tdhock/change-tutorial"
## [2] "https://rcdata.nau.edu/genomic-ml/animint-gallery/2016-01-28-Max-margin-interval-regression-for-supervised-segmentation-model-selection/index.html"
## [3] "https://rcdata.nau.edu/genomic-ml/animint-gallery/2016-11-10-Max-margin-supervised-penalty-learning-for-peak-detection-in-ChIP-seq-data/index.html"
##
## $`/home/local/USHERBROOKE/hoct2726/R/animint-manual-en/chapters/ch19/index.qmd`
## [1] "https://en.wikipedia.org/wiki/P-value"
## [2] "https://altair-viz.github.io/user_guide/large_datasets.html#preaggregate-and-filter-in-pandas"
## [3] "https://web.archive.org/web/20250827141658/https://plotly-r.com/performance"
##
## $`/home/local/USHERBROOKE/hoct2726/R/animint-manual-en/chapters/ch20/index.qmd`
## [1] "https://rdrr.io/cran/mlbench/man/Sonar.html"
The goal of this post is to write a conversion to bibtex.
The source code to find is please consult our [wiki FAQ](https://github.com/tdhock/animint2/wiki/FAQ#web-browser-on-local-indexhtml-file-is-blank).
We want to change it to
please consult our wiki FAQ [@animintFAQ]
@unpublished{animintFAQ,
title={Animint FAQ},
author={Toby Hocking},
url={https://github.com/tdhock/animint2/wiki/FAQ#web-browser-on-local-indexhtml-file-is-blank},
year={2025}
}
find in one file
Here is an example with one file.
qmd <- "~/R/animint-manual-en/chapters/ch14/index.qmd"
Below we define a regular expression to match a markdown link.
link_pattern <- list(
"\\[",
title=".*?",
"\\]\\(",
url="http.*?",
"\\)")
nc::capture_all_str(qmd, link_pattern)
## title
## <char>
## 1: PeakSegJoint package
## 2: this visualization of linear model and nearest neighbors
## url
## <char>
## 1: https://github.com/tdhock/PeakSegJoint
## 2: https://tdhock.github.io/2023-12-04-degree-neighbors/
The output above is a table with two lines, one for each markdown link in the qmd file.
This pattern is not sufficient for our purpose, which is to change each link to a citation.
To do that we need a pattern which will result in a set of matches that spans the whole file.
We do that below by adding
- a
beforegroup that captures anything before the link, even newlines, as indicated by(?s). - an alternative that matches the end of the file,
$, instead of the link.
before_pattern <- list(
before="(?s).+?",
nc::alternatives(
link=link_pattern,
"$"))
match_dt <- nc::capture_all_str(qmd, before_pattern)
tibble::tibble(match_dt)
## # A tibble: 3 × 4
## before link title url
## <chr> <chr> <chr> <chr>
## 1 "# Named `clickSelects` and `showSelected`\n\n```{r setup, echo=FALSE}\nknitr:… "[Pe… "Pea… "htt…
## 2 ", which is for peak detection in genomic data sequences.\nThe code below down… "[th… "thi… "htt…
## 3 ".\n* Use named `clickSelects` and `showSelected` to create a visualization of… "" "" ""
tibble::tibble(match_dt[, .(title, url)])
## # A tibble: 3 × 2
## title url
## <chr> <chr>
## 1 "PeakSegJoint package" "https://github.com/tdhock/PeakSegJoi…
## 2 "this visualization of linear model and nearest neighbors" "https://tdhock.github.io/2023-12-04-…
## 3 "" ""
The output above shows that there were three matches.
- The first two are links.
- The last one contains the rest of the file, after the last link.
Does this set of matches really span the whole file? Below we paste together the matched data, write it to a reconstituted file, and then read it back into R, along with the original file.
same.qmd <- tempfile()
same_content <- match_dt[, paste(paste0(before, link), collapse="")]
writeLines(same_content, same.qmd)
qlist <- list(original=qmd, reconstituted=same.qmd)
q_data_list <- sapply(qlist, readLines, simplify=FALSE)
with(q_data_list, identical(original, reconstituted))
## [1] TRUE
The output above indicates that we have indeed matched the whole file.
replace in one file
Below we compute the replacement citation for each markdown link.
match_dt[
, key := gsub(" ", "_", title)
][
, replacement := ifelse(
link=="", "",
sprintf("%s [@%s]", title, key))
][, replacement]
## [1] "PeakSegJoint package [@PeakSegJoint_package]"
## [2] "this visualization of linear model and nearest neighbors [@this_visualization_of_linear_model_and_nearest_neighbors]"
## [3] ""
We see above that title has been used to create the bibtex citation key which appears in square brackets.
Next, we verify that there are no violations using this replacement.
new_content <- match_dt[, paste(paste0(before, replacement), collapse="")]
get_violations(new_content)
## character(0)
get_violations(same_content)
## [1] "https://github.com/tdhock/PeakSegJoint"
## [2] "https://tdhock.github.io/2023-12-04-degree-neighbors/"
Above we see that there are no violations using the new content, but there were two violations using the old content. Finally we create the entries for the bib file.
bib_vec <- match_dt[-.N, sprintf("@unpublished{ %s,
title={ %s },
author={ TODO },
url={ %s },
year={ 2026 }
}
", key, title, url)]
cat(paste(bib_vec, collapse="\n"))
## @unpublished{ PeakSegJoint_package,
## title={ PeakSegJoint package },
## author={ TODO },
## url={ https://github.com/tdhock/PeakSegJoint },
## year={ 2026 }
## }
##
## @unpublished{ this_visualization_of_linear_model_and_nearest_neighbors,
## title={ this visualization of linear model and nearest neighbors },
## author={ TODO },
## url={ https://tdhock.github.io/2023-12-04-degree-neighbors/ },
## year={ 2026 }
## }
The output above shows two entries for a bib file.
find and replace in all files
chapters <- "~/R/animint-manual-en/chapters"
qmd.files <- c(
Sys.glob(file.path(chapters,"*qmd")),
Sys.glob(file.path(chapters,"*/index.qmd")))
bib_list <- list()
for(qmd in qmd.files){
match_dt <- nc::capture_all_str(qmd, before_pattern)[
, key := gsub(" ", "_", title)
][
, replacement := ifelse(
link=="", "",
sprintf("%s [@%s]", title, key))
]
new_content <- match_dt[, paste(paste0(before, replacement), collapse="")]
cat(new_content, file=qmd)
bib_list[[qmd]] <- match_dt[-.N, sprintf("@unpublished{ %s,
title={ %s },
author={ TODO },
url={ %s },
year={ 2026 }
}
", key, title, url)]
}
cat(paste(unlist(bib_list), collapse="\n"))
## @unpublished{ report_an_issue,
## title={ report an issue },
## author={ TODO },
## url={ https://github.com/animint/animint-manual-en/issues/new },
## year={ 2026 }
## }
##
## @unpublished{ the_famous_tea_tasting_experiment_proposed_by_Ronald_Fisher,
## title={ the famous tea tasting experiment proposed by Ronald Fisher },
## author={ TODO },
## url={ https://en.wikipedia.org/wiki/Lady_tasting_tea },
## year={ 2026 }
## }
##
## @unpublished{ Keeling_Curve,
## title={ Keeling Curve },
## author={ TODO },
## url={ https://en.wikipedia.org/wiki/Keeling_Curve },
## year={ 2026 }
## }
##
## @unpublished{ wiktionary,
## title={ wiktionary },
## author={ TODO },
## url={ https://en.wiktionary.org/wiki/sketch },
## year={ 2026 }
## }
##
## @unpublished{ S3_object_system,
## title={ S3 object system },
## author={ TODO },
## url={ http://adv-r.had.co.nz/OO-essentials.html#s3 },
## year={ 2026 }
## }
##
## @unpublished{ wiki_FAQ,
## title={ wiki FAQ },
## author={ TODO },
## url={ https://github.com/tdhock/animint2/wiki/FAQ#web-browser-on-local-indexhtml-file-is-blank },
## year={ 2026 }
## }
##
## @unpublished{ animation_package_examples,
## title={ animation package examples },
## author={ TODO },
## url={ https://yihui.name/animation/examples/ },
## year={ 2026 }
## }
##
## @unpublished{ R_Markdown,
## title={ R Markdown },
## author={ TODO },
## url={ http://rmarkdown.rstudio.com/ },
## year={ 2026 }
## }
##
## @unpublished{ GitHub_pages,
## title={ GitHub pages },
## author={ TODO },
## url={ https://pages.github.com/ },
## year={ 2026 }
## }
##
## @unpublished{ Netlify_Drop,
## title={ Netlify Drop },
## author={ TODO },
## url={ https://app.netlify.com/drop },
## year={ 2026 }
## }
##
## @unpublished{ the_Deploy_Overview_docs,
## title={ the Deploy Overview docs },
## author={ TODO },
## url={ https://docs.netlify.com/deploy/deploy-overview/ },
## year={ 2026 }
## }
##
## @unpublished{ GitHub_Pages,
## title={ GitHub Pages },
## author={ TODO },
## url={ https://pages.github.com/ },
## year={ 2026 }
## }
##
## @unpublished{ tdhock/cs499-spring2020,
## title={ tdhock/cs499-spring2020 },
## author={ TODO },
## url={ https://github.com/tdhock/cs499-spring2020 },
## year={ 2026 }
## }
##
## @unpublished{ figure-quadratic-interactive.R,
## title={ figure-quadratic-interactive.R },
## author={ TODO },
## url={ https://github.com/tdhock/cs499-spring2020/blob/master/2020-02-03-capacity/figure-quadratic-interactive.R },
## year={ 2026 }
## }
##
## @unpublished{ vimeo,
## title={ vimeo },
## author={ TODO },
## url={ https://vimeo.com },
## year={ 2026 }
## }
##
## @unpublished{ figure-several-interactive.R,
## title={ figure-several-interactive.R },
## author={ TODO },
## url={ https://github.com/tdhock/cs499-spring2020/blob/master/2020-02-03-capacity/figure-several-interactive.R },
## year={ 2026 }
## }
##
## @unpublished{ figure-quadratic-interactive.R,
## title={ figure-quadratic-interactive.R },
## author={ TODO },
## url={ https://github.com/tdhock/cs499-spring2020/blob/master/2020-02-03-capacity/figure-quadratic-interactive.R },
## year={ 2026 }
## }
##
## @unpublished{ README.md,
## title={ README.md },
## author={ TODO },
## url={ https://github.com/tdhock/2020-02-03-capacity-polynomial-degree/blob/gh-pages/README.md },
## year={ 2026 }
## }
##
## @unpublished{ repos.txt,
## title={ repos.txt },
## author={ TODO },
## url={ https://github.com/animint/gallery/blob/gh-pages/repos.txt },
## year={ 2026 }
## }
##
## @unpublished{ SVG,
## title={ SVG },
## author={ TODO },
## url={ http://www.w3schools.com/html/html5_svg.asp },
## year={ 2026 }
## }
##
## @unpublished{ HTML_id_attribute,
## title={ HTML id attribute },
## author={ TODO },
## url={ http://www.w3schools.com/tags/att_global_id.asp },
## year={ 2026 }
## }
##
## @unpublished{ for_testing_the_animint_JavaScript_renderer_code,
## title={ for testing the animint JavaScript renderer code },
## author={ TODO },
## url={ https://github.com/tdhock/animint/wiki/Testing },
## year={ 2026 }
## }
##
## @unpublished{ WorldBank_data_visualization_with_two_time_series_facets,
## title={ WorldBank data visualization with two time series facets },
## author={ TODO },
## url={ https://tdhock.github.io/2025-01-WorldBank-facets-map/ },
## year={ 2026 }
## }
##
## @unpublished{ WorldBank_data_visualization,
## title={ WorldBank data visualization },
## author={ TODO },
## url={ https://tdhock.github.io/2025-01-WorldBank-facets-map/ },
## year={ 2026 }
## }
##
## @unpublished{ this_visualization_of_cross-validation_for_change-point_model_selection,
## title={ this visualization of cross-validation for change-point model selection },
## author={ TODO },
## url={ https://tdhock.github.io/figure-binseg-cv-most-frequently-selected-fr/ },
## year={ 2026 }
## }
##
## @unpublished{ selectize.js,
## title={ selectize.js },
## author={ TODO },
## url={ http://selectize.github.io/selectize.js/ },
## year={ 2026 }
## }
##
## @unpublished{ PredictedPeaks_test_in_the_`animint2`_source_code,
## title={ PredictedPeaks test in the `animint2` source code },
## author={ TODO },
## url={ https://github.com/tdhock/animint2/blob/master/tests/testthat/test-renderer2-PredictedPeaks.R },
## year={ 2026 }
## }
##
## @unpublished{ Pull_Request,
## title={ Pull Request },
## author={ TODO },
## url={ https://github.com/tdhock/animint2/compare },
## year={ 2026 }
## }
##
## @unpublished{ issue_explaining_how_to_implement_it,
## title={ issue explaining how to implement it },
## author={ TODO },
## url={ https://github.com/tdhock/animint/issues/148 },
## year={ 2026 }
## }
##
## @unpublished{ an_issue,
## title={ an issue },
## author={ TODO },
## url={ https://github.com/tdhock/animint/issues/149 },
## year={ 2026 }
## }
##
## @unpublished{ is_an_issue,
## title={ is an issue },
## author={ TODO },
## url={ https://github.com/animint/animint2/issues/230 },
## year={ 2026 }
## }
##
## @unpublished{ all_of_the_theme_options,
## title={ all of the theme options },
## author={ TODO },
## url={ http://docs.ggplot2.org/current/theme.html },
## year={ 2026 }
## }
##
## @unpublished{ this_gallery_example,
## title={ this gallery example },
## author={ TODO },
## url={ https://tdhock.github.io/2025-01-WorldBank-facets-map/ },
## year={ 2026 }
## }
##
## @unpublished{ Elements_of_Statistical_Learning,
## title={ Elements of Statistical Learning },
## author={ TODO },
## url={ http://statweb.stanford.edu/~tibs/ElemStatLearn/ },
## year={ 2026 }
## }
##
## @unpublished{ Lasso,
## title={ Lasso },
## author={ TODO },
## url={ https://en.wikipedia.org/wiki/Lasso_%28statistics%29 },
## year={ 2026 }
## }
##
## @unpublished{ Support_Vector_Machine,
## title={ Support Vector Machine },
## author={ TODO },
## url={ https://en.wikipedia.org/wiki/Support_vector_machine },
## year={ 2026 }
## }
##
## @unpublished{ Poisson_regression,
## title={ Poisson regression },
## author={ TODO },
## url={ https://en.wikipedia.org/wiki/Poisson_regression },
## year={ 2026 }
## }
##
## @unpublished{ Binomial_regression,
## title={ Binomial regression },
## author={ TODO },
## url={ https://en.wikipedia.org/wiki/Binomial_regression },
## year={ 2026 }
## }
##
## @unpublished{ PeakSegJoint_package,
## title={ PeakSegJoint package },
## author={ TODO },
## url={ https://github.com/tdhock/PeakSegJoint },
## year={ 2026 }
## }
##
## @unpublished{ this_visualization_of_linear_model_and_nearest_neighbors,
## title={ this visualization of linear model and nearest neighbors },
## author={ TODO },
## url={ https://tdhock.github.io/2023-12-04-degree-neighbors/ },
## year={ 2026 }
## }
##
## @unpublished{ Lambert_W_function,
## title={ Lambert W function },
## author={ TODO },
## url={ https://www.wolframalpha.com/input/?i=a*x+%2Bb*log%28x%29%2B+c%3D0 },
## year={ 2026 }
## }
##
## @unpublished{ Newton's_method,
## title={ Newton's method },
## author={ TODO },
## url={ https://en.wikipedia.org/wiki/Newton%27s_method },
## year={ 2026 }
## }
##
## @unpublished{ Binomial_regression,
## title={ Binomial regression },
## author={ TODO },
## url={ https://en.wikipedia.org/wiki/Binomial_regression },
## year={ 2026 }
## }
##
## @unpublished{ my_useR_2017_tutorial,
## title={ my useR 2017 tutorial },
## author={ TODO },
## url={ https://github.com/tdhock/change-tutorial },
## year={ 2026 }
## }
##
## @unpublished{ like_this,
## title={ like this },
## author={ TODO },
## url={ https://rcdata.nau.edu/genomic-ml/animint-gallery/2016-01-28-Max-margin-interval-regression-for-supervised-segmentation-model-selection/index.html },
## year={ 2026 }
## }
##
## @unpublished{ like_this,
## title={ like this },
## author={ TODO },
## url={ https://rcdata.nau.edu/genomic-ml/animint-gallery/2016-11-10-Max-margin-supervised-penalty-learning-for-peak-detection-in-ChIP-seq-data/index.html },
## year={ 2026 }
## }
##
## @unpublished{ P-values,
## title={ P-values },
## author={ TODO },
## url={ https://en.wikipedia.org/wiki/P-value },
## year={ 2026 }
## }
##
## @unpublished{ altair,
## title={ altair },
## author={ TODO },
## url={ https://altair-viz.github.io/user_guide/large_datasets.html#preaggregate-and-filter-in-pandas },
## year={ 2026 }
## }
##
## @unpublished{ plotly,
## title={ plotly },
## author={ TODO },
## url={ https://web.archive.org/web/20250827141658/https://plotly-r.com/performance },
## year={ 2026 }
## }
##
## @unpublished{ `sonar`,
## title={ `sonar` },
## author={ TODO },
## url={ https://rdrr.io/cran/mlbench/man/Sonar.html },
## year={ 2026 }
## }
##
## @unpublished{ ,
## title={ },
## author={ TODO },
## url={ },
## year={ 2026 }
## }
After running the code above, I copied the output to the end of the refs.bib file, then manually corrected the TODOs in PR19.
Conclusion
We have seen how regex can be used for a non-trivial find and replace across multiple qmd files: changing markdown links to bib citations.
The before_pattern is generally useful for matching an entire file, so I created an issue in nc about potentially creating a pattern-making function.
Bonus: don’t use gsub etc.
You may be tempted to use gsub() or similar string replacement functions, but they actually do not work in this case.
gsub(
"\\[(.*?)\\]\\((http.*?)\\)",
"\\[@\\1\\]",
"[foo and](http) [bar not](http)",
perl=TRUE)
## [1] "[@foo and] [@bar not]"
The output above shows that the replacement works, but there are spaces in the bibtex citation keys, which are not allowed.
Note the \1 in the code above (numbered reference to capture group) can be replaced by a named reference ${title} using ICU in the code below.
subject <- "[foo](http) [bar](http)"
pattern <- "\\[(?<title>.*?)\\]\\((?<url>http.*?)\\)"
stringi::stri_replace_all_regex(subject, pattern, "\\[@${title}\\]")
## [1] "[@foo] [@bar]"
But surprisingly, this does not work with gsub() in the code below.
gsub(pattern, "\\[@${title}\\]", subject, perl=TRUE)
## [1] "[@${title}] [@${title}]"
gsub(pattern, "\\[@$<title>\\]", subject, perl=TRUE)
## [1] "[@$<title>] [@$<title>]"
gsub(pattern, "\\[@$1\\]", subject, perl=TRUE)
## [1] "[@$1] [@$1]"
gsub(pattern, "\\[@\\1\\]", subject, perl=TRUE)
## [1] "[@foo] [@bar]"
The only replacement syntax that works is \1 (number not name).
Is this a bug in R or PCRE2?
man pcre2syntaxsection REPLACEMENT STRINGS says we should be able to use$1for first group, and$<title>for named group.- R
?subargumentreplacementsays to use backslash, not dollar: For ‘fixed = FALSE’ this can include backreferences ‘”\1”’ to ‘”\9”’ to parenthesized subexpressions of ‘pattern’.
So R is working as documented.
But it is not clear to me why the R documentation differs from man pcre2syntax.
session info
sessionInfo()
## R Under development (unstable) (2026-02-07 r89380)
## Platform: x86_64-pc-linux-gnu
## Running under: Ubuntu 24.04.3 LTS
##
## Matrix products: default
## BLAS: /usr/lib/x86_64-linux-gnu/blas/libblas.so.3.12.0
## LAPACK: /usr/lib/x86_64-linux-gnu/lapack/liblapack.so.3.12.0 LAPACK version 3.12.0
##
## locale:
## [1] LC_CTYPE=fr_FR.UTF-8 LC_NUMERIC=C LC_TIME=fr_FR.UTF-8
## [4] LC_COLLATE=fr_FR.UTF-8 LC_MONETARY=fr_FR.UTF-8 LC_MESSAGES=fr_FR.UTF-8
## [7] LC_PAPER=fr_FR.UTF-8 LC_NAME=C LC_ADDRESS=C
## [10] LC_TELEPHONE=C LC_MEASUREMENT=fr_FR.UTF-8 LC_IDENTIFICATION=C
##
## time zone: America/Toronto
## tzcode source: system (glibc)
##
## attached base packages:
## [1] stats graphics grDevices utils datasets methods base
##
## other attached packages:
## [1] data.table_1.18.2.1
##
## loaded via a namespace (and not attached):
## [1] utf8_1.2.6 xfun_0.56 nc_2025.3.24 magrittr_2.0.4 glue_1.8.0
## [6] tibble_3.3.1 knitr_1.51 pkgconfig_2.0.3 lifecycle_1.0.5 cli_3.6.5
## [11] vctrs_0.7.1 compiler_4.6.0 tools_4.6.0 evaluate_1.0.5 pillar_1.11.1
## [16] otel_0.2.0 rlang_1.1.7 stringi_1.8.7