Volcano plots
The goal of this post is to describe a similarity between volcano plots and the SOAK summary plots that appear in our recent paper, SOAK: Same/Other/All K-fold cross-validation for estimating similarity of patterns in data subsets, arXiv:2410.08643.
Volcano plot
Volcano plots are a classic representation of effect size and statistical significance, when doing differential expression analysis in genomic data. For example, the sanssouci github repo has the volcano plot below:
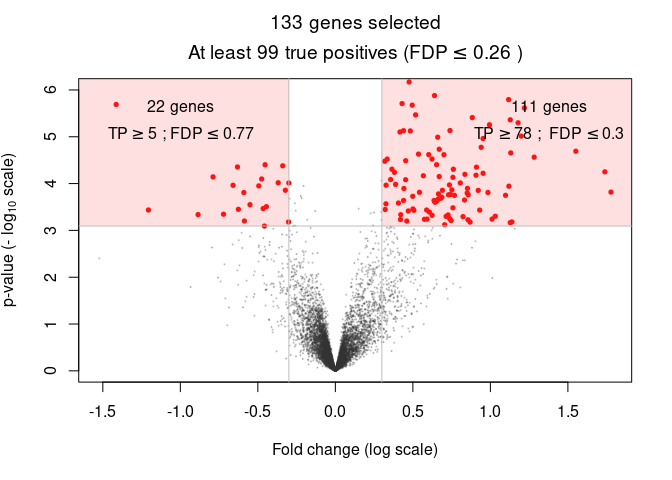
Each dot in the volcano plot represents a different gene. The genes which appear at the bottom center have little difference between groups. The genes which appear in the upper left or right have significant differences between groups.
SOAK plot
The slides for presenting my SOAK paper have the plot below:
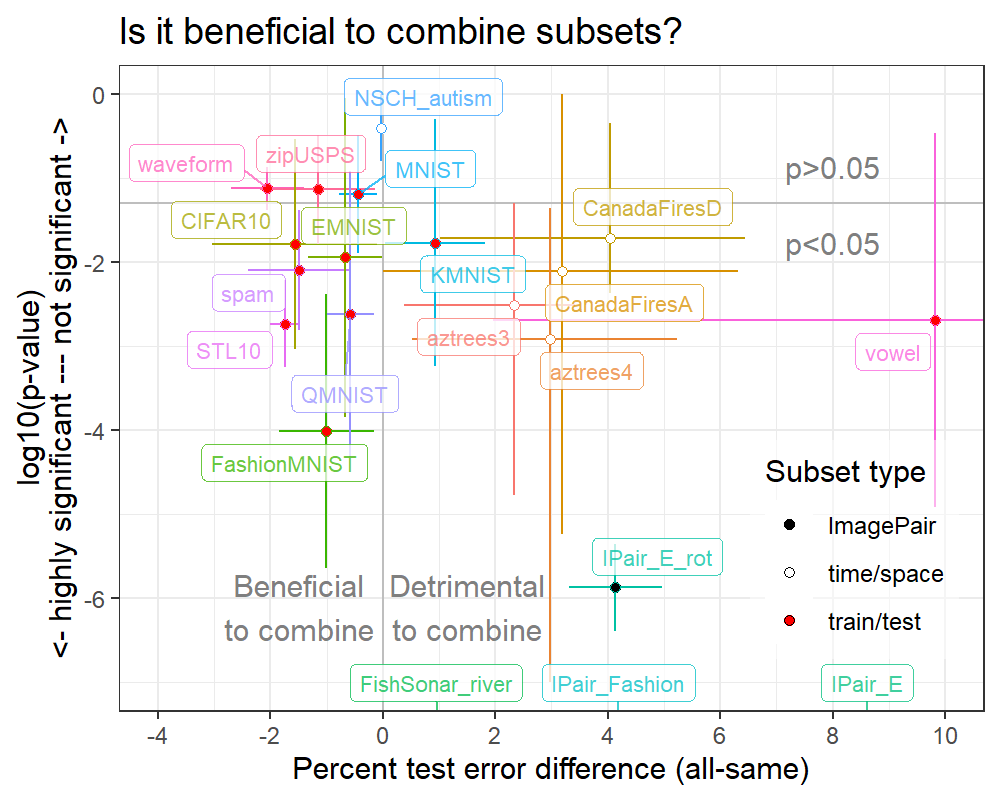
Each dot in the plot above represents one of twenty data sets that we analyzed using SOAK. The line segments represent the range of differences and p-values observed across the 2-4 data subsets. The typical V shape is not apparent because:
- The Y axis in inverted with respect to the volcano plot, and
- The line segments are useful to summarize each data set, but hide the details about the value of each data subset.
Re-making the SOAK plot like a volcano plot
data.list <- list()
local.csv.vec <- c("data-meta.csv", "data_Classif_batchmark_registry.csv")
prefix <- "https://raw.githubusercontent.com/tdhock/cv-same-other-paper/refs/heads/main/"
for(local.csv in local.csv.vec){
if(!file.exists(local.csv)){
u <- paste0(prefix, local.csv)
download.file(u, local.csv)
}
data.list[[local.csv]] <- fread(local.csv)
}
library(data.table)
(score.atomic <- data.list$data_Class[, .(
data.name,
test_subset = test.group,
train_subsets = train.groups,
test.fold, task_id, algorithm, percent.error
)])
## data.name test_subset train_subsets test.fold task_id algorithm percent.error
## <char> <char> <char> <int> <char> <char> <num>
## 1: aztrees3 NE all 1 aztrees3 cv_glmnet 0.000000
## 2: aztrees3 NW all 1 aztrees3 cv_glmnet 1.910828
## 3: aztrees3 S all 1 aztrees3 cv_glmnet 4.095563
## 4: aztrees3 NE all 2 aztrees3 cv_glmnet 4.081633
## 5: aztrees3 NW all 2 aztrees3 cv_glmnet 1.910828
## ---
## 2932: zipUSPS train same 8 zipUSPS featureless 83.608815
## 2933: zipUSPS test same 9 zipUSPS featureless 81.725888
## 2934: zipUSPS train same 9 zipUSPS featureless 83.586207
## 2935: zipUSPS test same 10 zipUSPS featureless 82.142857
## 2936: zipUSPS train same 10 zipUSPS featureless 83.586207
The table above contains the results of running SOAK with two
algorithms (cv_glmnet and featureless) on 20 classification data
sets. The percent.error column contains the test error for
predicting on test_subset, after training on train_subsets.
Data <- function(data.name, Data){
data.table(data.name, Data)
}
disp.dt <- rbind(
Data("CanadaFires_all","CanadaFiresA"),
Data("CanadaFires_downSampled","CanadaFiresD"),
Data("MNIST_EMNIST", "IPair_E"),
Data("MNIST_EMNIST_rot", "IPair_E_rot"),
Data("MNIST_FashionMNIST","IPair_Fashion"))
meta.raw <- data.list[["data-meta.csv"]][
grepl("train|test",group.small.name), `:=`(
test=ifelse(group.small.name=="test", group.small.N, group.large.N),
train=ifelse(group.small.name=="train", group.small.N, group.large.N)
)
][
, `test%` := as.integer(100*test/rows)
][
, subset_type := fcase(
grepl("MNIST_", data.name), "ImagePair",
!is.na(test), "train/test",
default="time/space")
][]
meta.dt <- disp.dt[
meta.raw, on="data.name"
][is.na(Data), Data := data.name][]
meta.dt[order(subset_type,data.name), .(subset_type, data.name, group.tab)]
## subset_type data.name group.tab
## <char> <char> <char>
## 1: ImagePair MNIST_EMNIST EMNIST=70000;MNIST=70000
## 2: ImagePair MNIST_EMNIST_rot EMNIST_rot=70000;MNIST=70000
## 3: ImagePair MNIST_FashionMNIST FashionMNIST=70000;MNIST=70000
## 4: time/space CanadaFires_all 306=364;329=755;395=1170;326=2538
## 5: time/space CanadaFires_downSampled 306=287;329=304;326=450;395=450
## 6: time/space FishSonar_river CHI=638475;PRL=667201;LEA=739745;BOU=770323
## 7: time/space NSCH_autism 2019=18202;2020=27808
## 8: time/space aztrees3 NE=1464;NW=1563;S=2929
## 9: time/space aztrees4 SW=497;NE=1464;NW=1563;SE=2432
## 10: train/test CIFAR10 test=10000;train=50000
## 11: train/test EMNIST test=10000;train=60000
## 12: train/test FashionMNIST test=10000;train=60000
## 13: train/test KMNIST test=10000;train=60000
## 14: train/test MNIST test=10000;train=60000
## 15: train/test QMNIST test=60000;train=60000
## 16: train/test STL10 train=5000;test=8000
## 17: train/test spam test=1536;train=3065
## 18: train/test vowel test=462;train=528
## 19: train/test waveform train=300;test=500
## 20: train/test zipUSPS test=2007;train=7291
## subset_type data.name group.tab
The table above contains meta-data, one row for each of the twenty data sets analyzed using SOAK. Next, we compute a wide table of percent error values:
group.meta <- meta.dt[, nc::capture_all_str(
group.tab,
test_subset="[^;]+",
"=",
subset_rows="[0-9]+", as.integer
), by=data.name]
score.join <- meta.dt[score.atomic, on="data.name"]
(tab.wide <- dcast(
score.join[algorithm=="cv_glmnet"],
subset_type + Data + test_subset + test.fold ~ train_subsets,
value.var="percent.error"))
## Key: <subset_type, Data, test_subset, test.fold>
## subset_type Data test_subset test.fold all other same
## <char> <char> <char> <int> <num> <num> <num>
## 1: ImagePair IPair_E EMNIST 1 15.228571 81.900000 6.357143
## 2: ImagePair IPair_E EMNIST 2 15.914286 81.214286 6.657143
## 3: ImagePair IPair_E EMNIST 3 14.842857 81.042857 6.457143
## 4: ImagePair IPair_E EMNIST 4 15.442857 80.600000 6.542857
## 5: ImagePair IPair_E EMNIST 5 15.728571 80.057143 6.642857
## ---
## 486: train/test zipUSPS train 6 3.988996 6.740028 4.126547
## 487: train/test zipUSPS train 7 5.234160 7.988981 4.958678
## 488: train/test zipUSPS train 8 5.371901 7.851240 5.647383
## 489: train/test zipUSPS train 9 4.413793 8.275862 5.241379
## 490: train/test zipUSPS train 10 5.655172 7.862069 6.206897
The table above has one row for each combination of (Data set,
test.fold, test_subset), and one column for each value of
train_subsets (all, other, same). We convert to long form below:
(tab.long.raw <- melt(
tab.wide,
measure=c("other","all"),
variable.name="compare_name",
value.name="compare_error"))
## subset_type Data test_subset test.fold same compare_name compare_error
## <char> <char> <char> <int> <num> <fctr> <num>
## 1: ImagePair IPair_E EMNIST 1 6.357143 other 81.900000
## 2: ImagePair IPair_E EMNIST 2 6.657143 other 81.214286
## 3: ImagePair IPair_E EMNIST 3 6.457143 other 81.042857
## 4: ImagePair IPair_E EMNIST 4 6.542857 other 80.600000
## 5: ImagePair IPair_E EMNIST 5 6.642857 other 80.057143
## ---
## 976: train/test zipUSPS train 6 4.126547 all 3.988996
## 977: train/test zipUSPS train 7 4.958678 all 5.234160
## 978: train/test zipUSPS train 8 5.647383 all 5.371901
## 979: train/test zipUSPS train 9 5.241379 all 4.413793
## 980: train/test zipUSPS train 10 6.206897 all 5.655172
The long form above lets us compare same with other and all, using the P-value computation below:
computeP <- function(...){
by.vec <- c(...)
tab.long.raw[, {
test.res <- t.test(compare_error, same, paired=TRUE)
log.signed.p <- function(signed.p){
sign(signed.p)*abs(log10(abs(signed.p)))
}
with(test.res, data.table(
estimate,
p.value,
compare_mean=mean(compare_error,na.rm=TRUE),
same_mean=mean(same,na.rm=TRUE),
log10.p=log10(p.value),
sign.log.p=log.signed.p(p.value*sign(estimate)),
N=.N))
}, by=by.vec]
}
p.each.group <- computeP("subset_type","Data","compare_name","test_subset")
compare.wide.groups <- dcast(
p.each.group,
subset_type + Data ~ compare_name,
list(min, max, mean),
value.var=c("estimate","log10.p"))
tab.long <- computeP("subset_type","Data","compare_name")
(compare.wide <- dcast(
tab.long,
subset_type + Data ~ compare_name,
value.var=c("estimate","log10.p")
)[compare.wide.groups, on=.(subset_type,Data)])
## Key: <subset_type, Data>
## subset_type Data estimate_other estimate_all log10.p_other log10.p_all estimate_min_other
## <char> <char> <num> <num> <num> <num> <num>
## 1: ImagePair IPair_E 73.94278441 8.58414037 -36.12557898 -19.9571317 73.21985454
## 2: ImagePair IPair_E_rot 37.78295684 4.13574119 -28.05149106 -10.0273169 36.73305654
## 3: ImagePair IPair_Fashion 81.12217939 3.60643915 -26.29380602 -11.1659249 77.52428571
## 4: time/space CanadaFiresA 12.73119454 3.19346986 -12.15139280 -4.4954774 2.37373163
## 5: time/space CanadaFiresD 11.08297211 4.04950901 -10.55342170 -5.4355425 7.23655914
## 6: time/space FishSonar_river 2.14222066 1.67869144 -9.46757756 -7.1417503 0.50044287
## 7: time/space NSCH_autism 0.06869678 -0.03295173 -0.80281281 -0.5788356 -0.06038222
## 8: time/space aztrees3 12.75254188 2.33742877 -4.94136087 -3.9006761 3.32686381
## 9: time/space aztrees4 10.91479089 2.97026492 -6.01927681 -4.5729462 3.01043395
## 10: train/test CIFAR10 0.06900000 -1.57100000 -0.03427972 -2.5555501 -2.79000000
## 11: train/test EMNIST -0.22719298 -0.70789474 -0.28990449 -2.6439777 -1.49000000
## 12: train/test FashionMNIST 0.27250000 -1.00583333 -0.25506863 -3.8370923 -1.62000000
## 13: train/test KMNIST 5.73250000 0.93333333 -10.54167700 -2.6079393 4.14000000
## 14: train/test MNIST 0.40268174 -0.44128882 -0.71578296 -2.0236324 -0.64938271
## 15: train/test QMNIST 0.20330352 -0.58918097 -0.49318750 -3.6638006 -0.59667262
## 16: train/test STL10 0.15750000 -1.73750000 -0.10956553 -4.6478898 -1.06000000
## 17: train/test spam -0.68438147 -1.49749989 -0.80213878 -3.3172339 -1.43284293
## 18: train/test vowel 28.70454545 9.81818182 -9.42176606 -3.4375486 23.50000000
## 19: train/test waveform -0.60749910 -2.05518719 -0.26171299 -1.5446149 -2.04671858
## 20: train/test zipUSPS 0.23993120 -1.16190955 -0.14697286 -1.8158690 -2.08458852
## subset_type Data estimate_other estimate_all log10.p_other log10.p_all estimate_min_other
## estimate_min_all log10.p_min_other log10.p_min_all estimate_max_other estimate_max_all log10.p_max_other
## <num> <num> <num> <num> <num> <num>
## 1: 8.162724e+00 -19.1855428 -11.1852296 74.6657143 9.052381e+00 -18.34494658
## 2: 3.312752e+00 -15.2819271 -6.3918049 38.8328571 4.958730e+00 -15.12765365
## 3: 2.584307e+00 -21.6603229 -12.3831809 84.7200731 4.628571e+00 -17.76668547
## 4: -2.218278e-17 -6.7029138 -5.2462861 20.5797080 6.321321e+00 -2.42289502
## 5: 1.010753e+00 -3.6935764 -2.3633517 16.0000000 6.444444e+00 -2.52272197
## 6: 4.752551e-01 -14.6703314 -14.3648065 4.6164502 4.364652e+00 -8.14097009
## 7: -6.589182e-02 -2.8915032 -0.7920586 0.1977758 -1.163559e-05 -0.46373255
## 8: 3.833958e-01 -7.9297112 -4.7714958 30.5950051 3.419998e+00 -2.93905476
## 9: 5.124228e-01 -9.1721995 -7.0049919 29.7819402 5.224490e+00 -1.66564731
## 10: -3.040000e+00 -7.2308941 -3.0308816 2.9280000 -1.020000e-01 -3.14915524
## 11: -1.330000e+00 -6.3649830 -3.8468297 1.1759259 -1.666667e-02 -3.70550402
## 12: -1.850000e+00 -6.2626646 -5.6422445 2.1650000 -1.616667e-01 -4.45852315
## 13: 4.666667e-02 -10.7469910 -3.2389992 7.3250000 1.820000e+00 -5.70697352
## 14: -7.792532e-01 -3.4218128 -1.8923095 1.4547462 -1.033244e-01 -1.59415295
## 15: -1.028346e+00 -4.9293022 -4.3218119 1.0032797 -1.500164e-01 -3.31918513
## 16: -2.000000e+00 -1.6011453 -3.2481787 1.3750000 -1.475000e+00 -0.63846190
## 17: -2.408148e+00 -1.0340034 -2.8057248 0.0640800 -5.868522e-01 -0.04967194
## 18: 1.954545e+00 -5.4806885 -4.9159399 33.9090909 1.768182e+01 -4.79314130
## 19: -2.701891e+00 -0.6362025 -1.3740040 0.8317204 -1.408483e+00 -0.33892053
## 20: -2.185365e+00 -6.7564815 -1.7849263 2.5644509 -1.384545e-01 -1.76993887
## estimate_min_all log10.p_min_other log10.p_min_all estimate_max_other estimate_max_all log10.p_max_other
## log10.p_max_all estimate_mean_other estimate_mean_all log10.p_mean_other log10.p_mean_all
## <num> <num> <num> <num> <num>
## 1: -1.019233e+01 73.94278441 8.60755240 -18.7652447 -10.6887780
## 2: -5.363052e+00 37.78295684 4.13574119 -15.2047904 -5.8774285
## 3: -8.637530e+00 81.12217939 3.60643915 -19.7135042 -10.5103556
## 4: 0.000000e+00 12.73119454 3.19346986 -5.3215789 -2.1054884
## 5: -3.372540e-01 11.08297211 4.04950901 -3.0903031 -1.7121430
## 6: -8.575531e+00 2.14222066 1.67869144 -12.6392296 -11.3247505
## 7: -1.067407e-04 0.06869678 -0.03295173 -1.6776179 -0.3960826
## 8: -1.295590e+00 12.75254188 2.33742877 -5.5645877 -2.5108077
## 9: -1.352162e+00 10.91479089 2.97026492 -4.8362095 -2.9181016
## 10: -5.324016e-01 0.06900000 -1.57100000 -5.1900247 -1.7816416
## 11: -4.231388e-02 -0.15703704 -0.67333333 -5.0352435 -1.9445718
## 12: -2.384369e+00 0.27250000 -1.00583333 -5.3605939 -4.0133067
## 13: -2.984299e-01 5.73250000 0.93333333 -8.2269822 -1.7687145
## 14: -4.840571e-01 0.40268174 -0.44128882 -2.5079829 -1.1881833
## 15: -9.119973e-01 0.20330352 -0.58918097 -4.1242437 -2.6169046
## 16: -2.216951e+00 0.15750000 -1.73750000 -1.1198036 -2.7325648
## 17: -1.384153e+00 -0.68438147 -1.49749989 -0.5418377 -2.0949387
## 18: -4.648515e-01 28.70454545 9.81818182 -5.1369149 -2.6903957
## 19: -8.655658e-01 -0.60749910 -2.05518719 -0.4875615 -1.1197849
## 20: -4.823761e-01 0.23993120 -1.16190955 -4.2632102 -1.1336512
## log10.p_max_all estimate_mean_other estimate_mean_all log10.p_mean_other log10.p_mean_all
The table above has one row per data set, and columns for different statistics (min, max, mean) of P-value (log10.p) and test error difference (estimate). The code below reproduces the SOAK figure.
compare.wide[log10.p_all < -12, log10.p_all := -Inf][]
## Key: <subset_type, Data>
## subset_type Data estimate_other estimate_all log10.p_other log10.p_all estimate_min_other
## <char> <char> <num> <num> <num> <num> <num>
## 1: ImagePair IPair_E 73.94278441 8.58414037 -36.12557898 -Inf 73.21985454
## 2: ImagePair IPair_E_rot 37.78295684 4.13574119 -28.05149106 -10.0273169 36.73305654
## 3: ImagePair IPair_Fashion 81.12217939 3.60643915 -26.29380602 -11.1659249 77.52428571
## 4: time/space CanadaFiresA 12.73119454 3.19346986 -12.15139280 -4.4954774 2.37373163
## 5: time/space CanadaFiresD 11.08297211 4.04950901 -10.55342170 -5.4355425 7.23655914
## 6: time/space FishSonar_river 2.14222066 1.67869144 -9.46757756 -7.1417503 0.50044287
## 7: time/space NSCH_autism 0.06869678 -0.03295173 -0.80281281 -0.5788356 -0.06038222
## 8: time/space aztrees3 12.75254188 2.33742877 -4.94136087 -3.9006761 3.32686381
## 9: time/space aztrees4 10.91479089 2.97026492 -6.01927681 -4.5729462 3.01043395
## 10: train/test CIFAR10 0.06900000 -1.57100000 -0.03427972 -2.5555501 -2.79000000
## 11: train/test EMNIST -0.22719298 -0.70789474 -0.28990449 -2.6439777 -1.49000000
## 12: train/test FashionMNIST 0.27250000 -1.00583333 -0.25506863 -3.8370923 -1.62000000
## 13: train/test KMNIST 5.73250000 0.93333333 -10.54167700 -2.6079393 4.14000000
## 14: train/test MNIST 0.40268174 -0.44128882 -0.71578296 -2.0236324 -0.64938271
## 15: train/test QMNIST 0.20330352 -0.58918097 -0.49318750 -3.6638006 -0.59667262
## 16: train/test STL10 0.15750000 -1.73750000 -0.10956553 -4.6478898 -1.06000000
## 17: train/test spam -0.68438147 -1.49749989 -0.80213878 -3.3172339 -1.43284293
## 18: train/test vowel 28.70454545 9.81818182 -9.42176606 -3.4375486 23.50000000
## 19: train/test waveform -0.60749910 -2.05518719 -0.26171299 -1.5446149 -2.04671858
## 20: train/test zipUSPS 0.23993120 -1.16190955 -0.14697286 -1.8158690 -2.08458852
## subset_type Data estimate_other estimate_all log10.p_other log10.p_all estimate_min_other
## estimate_min_all log10.p_min_other log10.p_min_all estimate_max_other estimate_max_all log10.p_max_other
## <num> <num> <num> <num> <num> <num>
## 1: 8.162724e+00 -19.1855428 -11.1852296 74.6657143 9.052381e+00 -18.34494658
## 2: 3.312752e+00 -15.2819271 -6.3918049 38.8328571 4.958730e+00 -15.12765365
## 3: 2.584307e+00 -21.6603229 -12.3831809 84.7200731 4.628571e+00 -17.76668547
## 4: -2.218278e-17 -6.7029138 -5.2462861 20.5797080 6.321321e+00 -2.42289502
## 5: 1.010753e+00 -3.6935764 -2.3633517 16.0000000 6.444444e+00 -2.52272197
## 6: 4.752551e-01 -14.6703314 -14.3648065 4.6164502 4.364652e+00 -8.14097009
## 7: -6.589182e-02 -2.8915032 -0.7920586 0.1977758 -1.163559e-05 -0.46373255
## 8: 3.833958e-01 -7.9297112 -4.7714958 30.5950051 3.419998e+00 -2.93905476
## 9: 5.124228e-01 -9.1721995 -7.0049919 29.7819402 5.224490e+00 -1.66564731
## 10: -3.040000e+00 -7.2308941 -3.0308816 2.9280000 -1.020000e-01 -3.14915524
## 11: -1.330000e+00 -6.3649830 -3.8468297 1.1759259 -1.666667e-02 -3.70550402
## 12: -1.850000e+00 -6.2626646 -5.6422445 2.1650000 -1.616667e-01 -4.45852315
## 13: 4.666667e-02 -10.7469910 -3.2389992 7.3250000 1.820000e+00 -5.70697352
## 14: -7.792532e-01 -3.4218128 -1.8923095 1.4547462 -1.033244e-01 -1.59415295
## 15: -1.028346e+00 -4.9293022 -4.3218119 1.0032797 -1.500164e-01 -3.31918513
## 16: -2.000000e+00 -1.6011453 -3.2481787 1.3750000 -1.475000e+00 -0.63846190
## 17: -2.408148e+00 -1.0340034 -2.8057248 0.0640800 -5.868522e-01 -0.04967194
## 18: 1.954545e+00 -5.4806885 -4.9159399 33.9090909 1.768182e+01 -4.79314130
## 19: -2.701891e+00 -0.6362025 -1.3740040 0.8317204 -1.408483e+00 -0.33892053
## 20: -2.185365e+00 -6.7564815 -1.7849263 2.5644509 -1.384545e-01 -1.76993887
## estimate_min_all log10.p_min_other log10.p_min_all estimate_max_other estimate_max_all log10.p_max_other
## log10.p_max_all estimate_mean_other estimate_mean_all log10.p_mean_other log10.p_mean_all
## <num> <num> <num> <num> <num>
## 1: -1.019233e+01 73.94278441 8.60755240 -18.7652447 -10.6887780
## 2: -5.363052e+00 37.78295684 4.13574119 -15.2047904 -5.8774285
## 3: -8.637530e+00 81.12217939 3.60643915 -19.7135042 -10.5103556
## 4: 0.000000e+00 12.73119454 3.19346986 -5.3215789 -2.1054884
## 5: -3.372540e-01 11.08297211 4.04950901 -3.0903031 -1.7121430
## 6: -8.575531e+00 2.14222066 1.67869144 -12.6392296 -11.3247505
## 7: -1.067407e-04 0.06869678 -0.03295173 -1.6776179 -0.3960826
## 8: -1.295590e+00 12.75254188 2.33742877 -5.5645877 -2.5108077
## 9: -1.352162e+00 10.91479089 2.97026492 -4.8362095 -2.9181016
## 10: -5.324016e-01 0.06900000 -1.57100000 -5.1900247 -1.7816416
## 11: -4.231388e-02 -0.15703704 -0.67333333 -5.0352435 -1.9445718
## 12: -2.384369e+00 0.27250000 -1.00583333 -5.3605939 -4.0133067
## 13: -2.984299e-01 5.73250000 0.93333333 -8.2269822 -1.7687145
## 14: -4.840571e-01 0.40268174 -0.44128882 -2.5079829 -1.1881833
## 15: -9.119973e-01 0.20330352 -0.58918097 -4.1242437 -2.6169046
## 16: -2.216951e+00 0.15750000 -1.73750000 -1.1198036 -2.7325648
## 17: -1.384153e+00 -0.68438147 -1.49749989 -0.5418377 -2.0949387
## 18: -4.648515e-01 28.70454545 9.81818182 -5.1369149 -2.6903957
## 19: -8.655658e-01 -0.60749910 -2.05518719 -0.4875615 -1.1197849
## 20: -4.823761e-01 0.23993120 -1.16190955 -4.2632102 -1.1336512
## log10.p_max_all estimate_mean_other estimate_mean_all log10.p_mean_other log10.p_mean_all
compare.wide[log10.p_other < -20, log10.p_other := -Inf][]
## Key: <subset_type, Data>
## subset_type Data estimate_other estimate_all log10.p_other log10.p_all estimate_min_other
## <char> <char> <num> <num> <num> <num> <num>
## 1: ImagePair IPair_E 73.94278441 8.58414037 -Inf -Inf 73.21985454
## 2: ImagePair IPair_E_rot 37.78295684 4.13574119 -Inf -10.0273169 36.73305654
## 3: ImagePair IPair_Fashion 81.12217939 3.60643915 -Inf -11.1659249 77.52428571
## 4: time/space CanadaFiresA 12.73119454 3.19346986 -12.15139280 -4.4954774 2.37373163
## 5: time/space CanadaFiresD 11.08297211 4.04950901 -10.55342170 -5.4355425 7.23655914
## 6: time/space FishSonar_river 2.14222066 1.67869144 -9.46757756 -7.1417503 0.50044287
## 7: time/space NSCH_autism 0.06869678 -0.03295173 -0.80281281 -0.5788356 -0.06038222
## 8: time/space aztrees3 12.75254188 2.33742877 -4.94136087 -3.9006761 3.32686381
## 9: time/space aztrees4 10.91479089 2.97026492 -6.01927681 -4.5729462 3.01043395
## 10: train/test CIFAR10 0.06900000 -1.57100000 -0.03427972 -2.5555501 -2.79000000
## 11: train/test EMNIST -0.22719298 -0.70789474 -0.28990449 -2.6439777 -1.49000000
## 12: train/test FashionMNIST 0.27250000 -1.00583333 -0.25506863 -3.8370923 -1.62000000
## 13: train/test KMNIST 5.73250000 0.93333333 -10.54167700 -2.6079393 4.14000000
## 14: train/test MNIST 0.40268174 -0.44128882 -0.71578296 -2.0236324 -0.64938271
## 15: train/test QMNIST 0.20330352 -0.58918097 -0.49318750 -3.6638006 -0.59667262
## 16: train/test STL10 0.15750000 -1.73750000 -0.10956553 -4.6478898 -1.06000000
## 17: train/test spam -0.68438147 -1.49749989 -0.80213878 -3.3172339 -1.43284293
## 18: train/test vowel 28.70454545 9.81818182 -9.42176606 -3.4375486 23.50000000
## 19: train/test waveform -0.60749910 -2.05518719 -0.26171299 -1.5446149 -2.04671858
## 20: train/test zipUSPS 0.23993120 -1.16190955 -0.14697286 -1.8158690 -2.08458852
## subset_type Data estimate_other estimate_all log10.p_other log10.p_all estimate_min_other
## estimate_min_all log10.p_min_other log10.p_min_all estimate_max_other estimate_max_all log10.p_max_other
## <num> <num> <num> <num> <num> <num>
## 1: 8.162724e+00 -19.1855428 -11.1852296 74.6657143 9.052381e+00 -18.34494658
## 2: 3.312752e+00 -15.2819271 -6.3918049 38.8328571 4.958730e+00 -15.12765365
## 3: 2.584307e+00 -21.6603229 -12.3831809 84.7200731 4.628571e+00 -17.76668547
## 4: -2.218278e-17 -6.7029138 -5.2462861 20.5797080 6.321321e+00 -2.42289502
## 5: 1.010753e+00 -3.6935764 -2.3633517 16.0000000 6.444444e+00 -2.52272197
## 6: 4.752551e-01 -14.6703314 -14.3648065 4.6164502 4.364652e+00 -8.14097009
## 7: -6.589182e-02 -2.8915032 -0.7920586 0.1977758 -1.163559e-05 -0.46373255
## 8: 3.833958e-01 -7.9297112 -4.7714958 30.5950051 3.419998e+00 -2.93905476
## 9: 5.124228e-01 -9.1721995 -7.0049919 29.7819402 5.224490e+00 -1.66564731
## 10: -3.040000e+00 -7.2308941 -3.0308816 2.9280000 -1.020000e-01 -3.14915524
## 11: -1.330000e+00 -6.3649830 -3.8468297 1.1759259 -1.666667e-02 -3.70550402
## 12: -1.850000e+00 -6.2626646 -5.6422445 2.1650000 -1.616667e-01 -4.45852315
## 13: 4.666667e-02 -10.7469910 -3.2389992 7.3250000 1.820000e+00 -5.70697352
## 14: -7.792532e-01 -3.4218128 -1.8923095 1.4547462 -1.033244e-01 -1.59415295
## 15: -1.028346e+00 -4.9293022 -4.3218119 1.0032797 -1.500164e-01 -3.31918513
## 16: -2.000000e+00 -1.6011453 -3.2481787 1.3750000 -1.475000e+00 -0.63846190
## 17: -2.408148e+00 -1.0340034 -2.8057248 0.0640800 -5.868522e-01 -0.04967194
## 18: 1.954545e+00 -5.4806885 -4.9159399 33.9090909 1.768182e+01 -4.79314130
## 19: -2.701891e+00 -0.6362025 -1.3740040 0.8317204 -1.408483e+00 -0.33892053
## 20: -2.185365e+00 -6.7564815 -1.7849263 2.5644509 -1.384545e-01 -1.76993887
## estimate_min_all log10.p_min_other log10.p_min_all estimate_max_other estimate_max_all log10.p_max_other
## log10.p_max_all estimate_mean_other estimate_mean_all log10.p_mean_other log10.p_mean_all
## <num> <num> <num> <num> <num>
## 1: -1.019233e+01 73.94278441 8.60755240 -18.7652447 -10.6887780
## 2: -5.363052e+00 37.78295684 4.13574119 -15.2047904 -5.8774285
## 3: -8.637530e+00 81.12217939 3.60643915 -19.7135042 -10.5103556
## 4: 0.000000e+00 12.73119454 3.19346986 -5.3215789 -2.1054884
## 5: -3.372540e-01 11.08297211 4.04950901 -3.0903031 -1.7121430
## 6: -8.575531e+00 2.14222066 1.67869144 -12.6392296 -11.3247505
## 7: -1.067407e-04 0.06869678 -0.03295173 -1.6776179 -0.3960826
## 8: -1.295590e+00 12.75254188 2.33742877 -5.5645877 -2.5108077
## 9: -1.352162e+00 10.91479089 2.97026492 -4.8362095 -2.9181016
## 10: -5.324016e-01 0.06900000 -1.57100000 -5.1900247 -1.7816416
## 11: -4.231388e-02 -0.15703704 -0.67333333 -5.0352435 -1.9445718
## 12: -2.384369e+00 0.27250000 -1.00583333 -5.3605939 -4.0133067
## 13: -2.984299e-01 5.73250000 0.93333333 -8.2269822 -1.7687145
## 14: -4.840571e-01 0.40268174 -0.44128882 -2.5079829 -1.1881833
## 15: -9.119973e-01 0.20330352 -0.58918097 -4.1242437 -2.6169046
## 16: -2.216951e+00 0.15750000 -1.73750000 -1.1198036 -2.7325648
## 17: -1.384153e+00 -0.68438147 -1.49749989 -0.5418377 -2.0949387
## 18: -4.648515e-01 28.70454545 9.81818182 -5.1369149 -2.6903957
## 19: -8.655658e-01 -0.60749910 -2.05518719 -0.4875615 -1.1197849
## 20: -4.823761e-01 0.23993120 -1.16190955 -4.2632102 -1.1336512
## log10.p_max_all estimate_mean_other estimate_mean_all log10.p_mean_other log10.p_mean_all
tlab <- function(x, y, label){
data.table(x, y, label)
}
text.y <- -6
text.dt <- rbind(
tlab(7, -1.7, "p<0.05"),
tlab(-2, text.y, "Beneficial\nto combine"),
tlab(8, text.y, "Detrimental\nto combine"))
set.seed(2)# for ggrepel.
type.colors <- c(
ImagePair="black",
"time/space"="white",
"train/test"="red")
library(ggplot2)
ggplot()+
ggtitle("Is it beneficial to combine subsets?")+
theme_bw()+
theme(legend.position=c(0.9,0.9))+
geom_hline(yintercept=log10(0.05),color="grey")+
geom_vline(xintercept=0,color="grey")+
geom_text(aes(
x, y, label=label, color=NULL),
color="grey50",
data=text.dt)+
geom_segment(aes(
estimate_min_all, log10.p_mean_all,
xend=estimate_max_all, yend=log10.p_mean_all,
color=Data),
data=compare.wide)+
geom_segment(aes(
estimate_mean_all, log10.p_min_all,
xend=estimate_mean_all, yend=log10.p_max_all,
color=Data),
data=compare.wide)+
geom_point(aes(
estimate_mean_all, log10.p_mean_all,
color=Data,
fill=subset_type),
shape=21,
data=compare.wide)+
ggrepel::geom_label_repel(aes(
estimate_mean_all, log10.p_mean_all, color=Data,
label=Data),
alpha=0.75,
size=2.8,
data=compare.wide)+
scale_fill_manual(
"Subset type", values=type.colors)+
scale_color_discrete(guide="none")+
scale_y_continuous(
"log10(p-value)\n<- highly significant --- not significant ->",
breaks=seq(-100,0,by=2))+
scale_x_continuous(
"Percent test error difference (all-same)",
breaks=seq(-100,10,by=2))+
coord_cartesian(
xlim=c(-4,10),
ylim=c(-7,0))
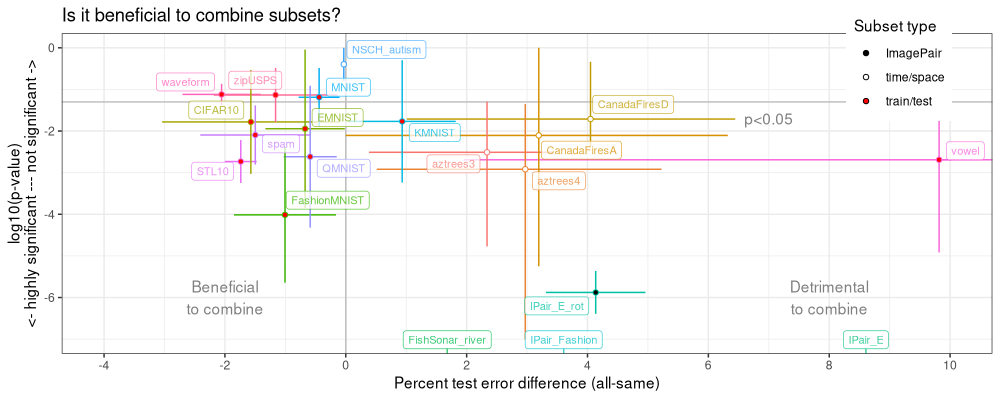
The SOAK figure above has one dot per data set, and segments which depict the range of P-values and test error differences, across the 2-4 data subsets. The values for each subset are in the table below:
(p.all <- p.each.group[compare_name=="all"])
## subset_type Data compare_name test_subset estimate p.value compare_mean same_mean
## <char> <char> <fctr> <char> <num> <num> <num> <num>
## 1: ImagePair IPair_E all EMNIST 9.052381e+00 6.527854e-12 15.479365 6.3771429
## 2: ImagePair IPair_E all MNIST 8.162724e+00 6.422049e-11 16.795679 8.6329553
## 3: ImagePair IPair_E_rot all EMNIST_rot 4.958730e+00 4.056908e-07 11.253968 6.3014286
## 4: ImagePair IPair_E_rot all MNIST 3.312752e+00 4.334588e-06 11.826428 8.5326816
## 5: ImagePair IPair_Fashion all FashionMNIST 4.628571e+00 4.138273e-13 19.117143 14.4885714
## 6: ImagePair IPair_Fashion all MNIST 2.584307e+00 2.303932e-09 11.028558 8.4442508
## 7: time/space CanadaFiresA all 306 6.321321e+00 8.250044e-03 10.180180 3.8588589
## 8: time/space CanadaFiresA all 326 4.736858e-01 8.088654e-02 4.768774 4.2950879
## 9: time/space CanadaFiresA all 329 -2.218278e-17 1.000000e+00 1.852726 1.8527264
## 10: time/space CanadaFiresA all 395 5.978872e+00 5.671709e-06 8.794452 2.8155799
## 11: time/space CanadaFiresD all 306 4.520617e+00 1.261188e-02 8.759807 4.2391899
## 12: time/space CanadaFiresD all 326 4.222222e+00 5.639676e-03 9.777778 5.5555556
## 13: time/space CanadaFiresD all 329 1.010753e+00 4.599875e-01 4.602151 3.5913978
## 14: time/space CanadaFiresD all 395 6.444444e+00 4.331599e-03 10.444444 4.0000000
## 15: time/space FishSonar_river all BOU 4.752551e-01 2.091857e-11 15.611503 15.1362481
## 16: time/space FishSonar_river all CHI 1.289792e+00 2.093166e-12 30.094209 28.8044168
## 17: time/space FishSonar_river all LEA 5.850666e-01 2.657473e-09 23.786981 23.2019142
## 18: time/space FishSonar_river all PRL 4.364652e+00 4.317113e-15 30.749504 26.3848521
## 19: time/space NSCH_autism all 2019 -6.589182e-02 1.614141e-01 2.527220 2.5931115
## 20: time/space NSCH_autism all 2020 -1.163559e-05 9.997543e-01 2.405782 2.4057932
## 21: time/space aztrees3 all NE 3.419998e+00 3.425018e-02 5.398379 1.9783804
## 22: time/space aztrees3 all NW 3.833958e-01 5.063021e-02 1.213882 0.8304858
## 23: time/space aztrees3 all S 3.208892e+00 1.692405e-05 3.379541 0.1706485
## 24: time/space aztrees4 all NE 3.677197e+00 4.444655e-02 6.408536 2.7313391
## 25: time/space aztrees4 all NW 5.124228e-01 3.669270e-02 1.342087 0.8296639
## 26: time/space aztrees4 all SE 2.466950e+00 9.885714e-08 2.466950 0.0000000
## 27: time/space aztrees4 all SW 5.224490e+00 1.318766e-02 5.224490 0.0000000
## 28: train/test CIFAR10 all test -3.040000e+00 9.313619e-04 58.600000 61.6400000
## 29: train/test CIFAR10 all train -1.020000e-01 2.934934e-01 58.746000 58.8480000
## 30: train/test EMNIST all test -1.330000e+00 1.422887e-04 6.420000 7.7500000
## 31: train/test EMNIST all train -1.666667e-02 9.071647e-01 6.410000 6.4759259
## 32: train/test FashionMNIST all test -1.850000e+00 2.279059e-06 15.490000 17.3400000
## 33: train/test FashionMNIST all train -1.616667e-01 4.126968e-03 14.273333 14.4350000
## 34: train/test KMNIST all test 1.820000e+00 5.767675e-04 28.050000 26.2300000
## 35: train/test KMNIST all train 4.666667e-02 5.030025e-01 17.846667 17.8000000
## 36: train/test MNIST all test -7.792532e-01 1.281417e-02 8.040451 8.8197042
## 37: train/test MNIST all train -1.033244e-01 3.280522e-01 8.606777 8.7101010
## 38: train/test QMNIST all test -1.500164e-01 1.224624e-01 7.521660 7.6716763
## 39: train/test QMNIST all train -1.028346e+00 4.766373e-05 7.599994 8.6283398
## 40: train/test STL10 all test -1.475000e+00 5.647046e-04 61.875000 63.3500000
## 41: train/test STL10 all train -2.000000e+00 6.068050e-03 62.920000 64.9200000
## 42: train/test spam all test -2.408148e+00 1.564139e-03 7.485434 9.8935821
## 43: train/test spam all train -5.868522e-01 4.129024e-02 7.799320 8.3861724
## 44: train/test vowel all test 1.768182e+01 1.213557e-05 34.409091 16.7272727
## 45: train/test vowel all train 1.954545e+00 3.428850e-01 32.045455 30.0909091
## 46: train/test waveform all test -1.408483e+00 4.226647e-02 15.177265 16.5857481
## 47: train/test waveform all train -2.701891e+00 1.362807e-01 17.315165 20.0170560
## 48: train/test zipUSPS all test -2.185365e+00 1.640868e-02 8.967641 11.1530060
## 49: train/test zipUSPS all train -1.384545e-01 3.293244e-01 5.045330 5.1837841
## subset_type Data compare_name test_subset estimate p.value compare_mean same_mean
## log10.p sign.log.p N
## <num> <num> <int>
## 1: -1.118523e+01 11.1852295929 10
## 2: -1.019233e+01 10.1923263848 10
## 3: -6.391805e+00 6.3918048928 10
## 4: -5.363052e+00 5.3630521512 10
## 5: -1.238318e+01 12.3831808513 10
## 6: -8.637530e+00 8.6375303401 10
## 7: -2.083544e+00 2.0835437513 10
## 8: -1.092124e+00 1.0921237608 10
## 9: 0.000000e+00 0.0000000000 10
## 10: -5.246286e+00 5.2462860843 10
## 11: -1.899220e+00 1.8992202560 10
## 12: -2.248746e+00 2.2487458549 10
## 13: -3.372540e-01 0.3372539975 10
## 14: -2.363352e+00 2.3633517073 10
## 15: -1.067947e+01 10.6794680920 10
## 16: -1.167920e+01 11.6791963545 10
## 17: -8.575531e+00 8.5755311130 10
## 18: -1.436481e+01 14.3648065475 10
## 19: -7.920586e-01 -0.7920585547 10
## 20: -1.067407e-04 -0.0001067407 10
## 21: -1.465337e+00 1.4653371418 10
## 22: -1.295590e+00 1.2955902359 10
## 23: -4.771496e+00 4.7714958171 10
## 24: -1.352162e+00 1.3521619376 10
## 25: -1.435420e+00 1.4354203344 10
## 26: -7.004992e+00 7.0049919447 10
## 27: -1.879832e+00 1.8798322378 10
## 28: -3.030882e+00 -3.0308815501 10
## 29: -5.324016e-01 -0.5324016450 10
## 30: -3.846830e+00 -3.8468296945 10
## 31: -4.231388e-02 -0.0423138804 10
## 32: -5.642244e+00 -5.6422444862 10
## 33: -2.384369e+00 -2.3843689300 10
## 34: -3.238999e+00 3.2389991852 10
## 35: -2.984299e-01 0.2984298594 10
## 36: -1.892309e+00 -1.8923094823 10
## 37: -4.840571e-01 -0.4840570812 10
## 38: -9.119973e-01 -0.9119972530 10
## 39: -4.321812e+00 -4.3218119439 10
## 40: -3.248179e+00 -3.2481786513 10
## 41: -2.216951e+00 -2.2169508763 10
## 42: -2.805725e+00 -2.8057247785 10
## 43: -1.384153e+00 -1.3841526090 10
## 44: -4.915940e+00 4.9159398671 10
## 45: -4.648515e-01 0.4648515495 10
## 46: -1.374004e+00 -1.3740040048 10
## 47: -8.655658e-01 -0.8655657936 10
## 48: -1.784926e+00 -1.7849263105 10
## 49: -4.823761e-01 -0.4823760539 10
## log10.p sign.log.p N
The table above has one row per subset in each of the data sets. To make a volcano plot, we need to invert the Y axis, and remove the segments.
text.y <- -6.5
text.dt <- rbind(
tlab(7, -1.4, "p<0.05"),
tlab(-2, text.y, "Beneficial\nto combine"),
tlab(8, text.y, "Detrimental\nto combine"))
ggplot()+
theme_bw()+
theme(legend.position=c(0.9,0.1))+
geom_hline(yintercept=-log10(0.05),color="grey")+
geom_vline(xintercept=0,color="grey")+
geom_text(aes(
x, -y, label=label, color=NULL),
color="grey50",
data=text.dt)+
geom_point(aes(
estimate, -log10.p,
color=Data,
fill=subset_type),
shape=21,
data=p.all)+
ggrepel::geom_label_repel(aes(
estimate, -log10.p, color=Data,
label=sprintf("%s\n%s",Data,test_subset)),
alpha=0.75,
size=2.8,
data=p.all)+
scale_fill_manual(
"Subset type", values=type.colors)+
scale_color_discrete(guide="none")+
scale_y_continuous(
"-log10(p-value)\n<- not significant --- highly significant ->",
breaks=seq(-100,100,by=2))+
scale_x_continuous(
"Percent test error difference (all-same)",
breaks=seq(-100,10,by=2))+
coord_cartesian(
xlim=c(-4,10),
ylim=c(0,7))
## Warning: ggrepel: 3 unlabeled data points (too many overlaps). Consider increasing max.overlaps
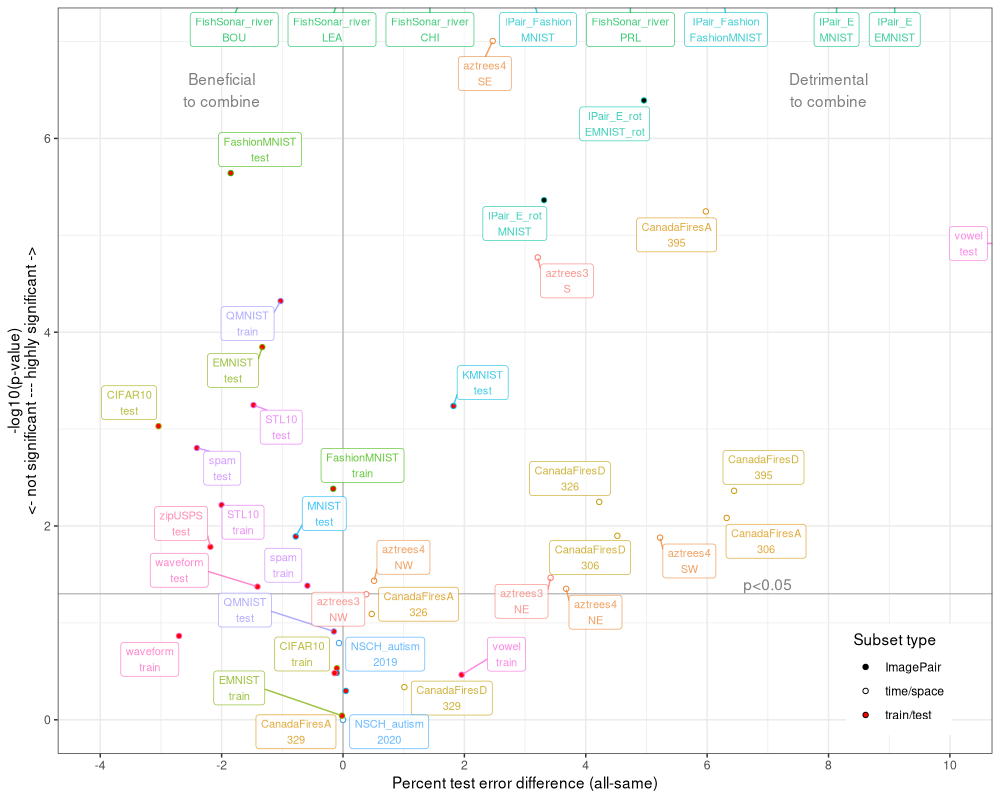
The result above has one dot (and label) for each data subset. The dots appear to have a distribution which is somewhat similar to the characteristic V pattern in volcano plots.
Conclusions
We have shown how to make volcano plots using SOAK subset data.
Session Info
sessionInfo()
## R version 4.5.0 (2025-04-11)
## Platform: x86_64-pc-linux-gnu
## Running under: Ubuntu 24.04.2 LTS
##
## Matrix products: default
## BLAS: /usr/lib/x86_64-linux-gnu/blas/libblas.so.3.12.0
## LAPACK: /usr/lib/x86_64-linux-gnu/lapack/liblapack.so.3.12.0 LAPACK version 3.12.0
##
## locale:
## [1] LC_CTYPE=fr_FR.UTF-8 LC_NUMERIC=C LC_TIME=fr_FR.UTF-8 LC_COLLATE=fr_FR.UTF-8
## [5] LC_MONETARY=fr_FR.UTF-8 LC_MESSAGES=fr_FR.UTF-8 LC_PAPER=fr_FR.UTF-8 LC_NAME=C
## [9] LC_ADDRESS=C LC_TELEPHONE=C LC_MEASUREMENT=fr_FR.UTF-8 LC_IDENTIFICATION=C
##
## time zone: Europe/Paris
## tzcode source: system (glibc)
##
## attached base packages:
## [1] stats graphics grDevices utils datasets methods base
##
## other attached packages:
## [1] ggplot2_3.5.1 data.table_1.17.0
##
## loaded via a namespace (and not attached):
## [1] directlabels_2024.1.21 crayon_1.5.3 vctrs_0.6.5 cli_3.6.4 knitr_1.50
## [6] xfun_0.51 rlang_1.1.5 ggrepel_0.9.6 bench_1.1.4 generics_0.1.3
## [11] glue_1.8.0 labeling_0.4.3 nc_2025.3.24 colorspace_2.1-1 scales_1.3.0
## [16] fpopw_1.1 quadprog_1.5-8 grid_4.5.0 evaluate_1.0.3 munsell_0.5.1
## [21] tibble_3.2.1 profmem_0.6.0 lifecycle_1.0.4 compiler_4.5.0 dplyr_1.1.4
## [26] Rcpp_1.0.14 pkgconfig_2.0.3 atime_2025.4.1 farver_2.1.2 lattice_0.22-6
## [31] R6_2.6.1 tidyselect_1.2.1 pillar_1.10.1 magrittr_2.0.3 tools_4.5.0
## [36] withr_3.0.2 gtable_0.3.6