History of supervised change-point detection
In my research about supervised change-point detection, I discovered a
link with previous work in the statistics literature: censored
regression, which is implemented in the R package survival. This post
is about how git bisect can be used to find a commit when survival
introduced a bug.
Background: history of supervised change-point detection
During my PHD, I was working at the Institute Curie, with medical doctors who were interested in classifying tumors based on their DNA copy number profiles. These data sets are like time series, but measured along space (positions on a chromosome), rather than time. If there are abrupt changes within a sequence, then the tumor is said to have “segmental copy number alternations,” and is likely to require more aggressive treatment (compared to a more benign sub-type in which these abrupt changes are not present).
Therefore, to accurately diagnose cancer patients, and propose appropriate treatments, it is important to accurately characterize the presence/absence of these abrupt changes. So I did a literature review of algorithms which could be used for detecting these change-points, and in my BMC Bioinformatics (2013) paper, we showed that the algorithms based on dynamic programming were most accurate, such as:
- The “segment neighborhood” method computes the best change-points
for a given number of segments. R implementations of this method can
be found in
jointseg::Fpsn,changepoint::cpt.mean(method="SegNeigh"), and (now archived) Segmentor3IsBack, cghseg::segmeanCO. - The “optimal partitioning” method computes the best change-points
for a given penalty, which is actually the same as the segment
neighborhood result, for some number of segments. R implementations
of this method can be found in
fpop::Fpop,changepoint::cpt.mean(method="PELT")and (now archived) gfpop.
To use these algorithms in practice for accurate detection, a model complexity parameter needs to be carefully chosen. That is either the number of segments (in the segment neighborhood method), or the penalty for each change (in the optimal partitioning method). The figure below, from my recent arXiv paper with my PHD student Tung Nguyen, is a very nice demonstration about how the penalty parameter affects the number of segments.

There are theoretical arguments that can be used to choose the model complexity parameter (AIC or BIC for example), in the unsupervised setting (no labels which indicate presence/absence of change-points in particular regions of data sequences). However, when there are labels available, our ICML’13 paper showed that it is much more accurate to use a supervised learning approach (where accuracy is measured with respect to labels drawn as colored rectangles in the figure above). The learning algorithm we proposed in that paper was called “max margin interval regression,” which uses gradient descent with a linear model and a squared hinge loss, where the label/output used in training is an interval of good penalty values for each labeled data sequence (right figures below). This is similar to usual regression which uses the square loss (left figures below, again from arXiv:2408.00856).


Our
algorithm is implemented in the penaltyLearning R package:
penaltyLearning::IntervalRegressionUnregularized (un-regularized)
and penaltyLearning::IntervalRegressionCV (degree of L1
regularization chosen using cross-validation).
The link with the statistics literature is that, in the context of
survival analysis, and in particular censored regression, the
label/output used in training can also be an interval. These
statistical models are called “Accelerated Failure Time” or AFT, and
are implemented as survival::survreg in R. Fitting the AFT Gaussian
model corresponds to minimizing a loss function with quadratic tails,
similar to our ICML’13 proposal based on the squared hinge loss.
With Rebecca Killick, we presented a tutorial at useR’17 about
change-point detection, including
figures
that show the similarity between these two kinds of loss
functions. With my GSOC’19 student Avinash Barnwal, and co-mentor
Philip Cho, we wrote a JCGS
paper
which described how the AFT loss functions could be used for censored
regression using the popular XGboost library.
Whereas survival::survreg is un-regularized, our proposal in ICML’13
was to use L1 regularization. I worked with several students to
implement iregnet, an R package
which attempts to implement AFT loss functions with L1
regularization. This package was meant to be analogous to glmnet, but
we never got the pruning rules to work as quickly as glmnet. I
suspect it would be better to implement L1-regularized AFT using the
new version of glmnet, which apparently can support all generalized
linear models, as described in the recent JSS
paper.
Other supervised change-point algorithms similar to our ICML’13 paper, which are worth mentioning here:
- Truong et al proposed an algorithm which learns based on labels that give the desired number of changes per sequence (rather than incomplete labels in regions), in EUSIPCO’17, but there is no R package that implements this method.
- At NeurIPS’17 we proposed Max Margin Interval Trees, which generalizes the classic greedy CART algorithm of Breiman, to the case of interval censored outputs. Code for R and Python on Github.
- Another supervised change-point algorithm that my student, Tung Nguyen, and I have been developing involves using a multi-layer perceptron model to predict the penalty value for the “optimal partitioning” algorithm: Deep Learning Approach for Changepoint Detection: Penalty Parameter Optimization. The figure below shows the proposed new idea relative to previous work, and the code is available on GitHub.

Issue
Back in 2017 when I was preparing the tutorial for useR, I ran into an issue, which I posted as
survival/issue#8 about survreg
not converging when all train labels are censored. In the case of my data, all outputs
are censored, which is normal for supervised change-point problems,
but rather unusual for survival analysis (where there is typically at
least one observation that is not censored). Because the “all
censored” train data is a rather niche use case for the survival AFT
model, I suppose it is not very well tested, so it is normal to expect
issues with such code.
After that issue was eventually fixed, survreg worked well enough so that I could use it to prepare my useR’17 tutorial on supervised change-point detection. Recently, a GSOC student tried reproducing some code from that time.
- The related issue, change-tutorial/issue#1 begins with Nhi observing the error “Supplied 2 items to be assigned to 1 items.”
- Using current R-4.4.1, I re-ran my old R code from 2017, which gave me warning “Ran out of iterations and did not converge” and error “pred must be a numeric vector or matrix with neither missing nor infinite entries” and saw that the problem was that survreg is returning missing values for the learned coefficients (where finite values were expected in the previous code).
- I made a copy of the R code, and edited it to make a minimal
reproducible example which demonstrates the bug.
That code runs survreg with three small data sets, each identified with a different value of
extra.id, and each with a few dozen data/rows. Running that code withsurvival_3.6-4I get the following results, for the data with:extra.id=0: NA coefs, convergence failed. Here I expected finite coefs, as I observed in 2017, after Terry fixed survival/issue#8.extra.id=13: finite coefs, converged. This was an expected result.extra.id=120: finite coefs, convergence failed. This was also unexpected, because it is inconsistent with the other two data sets above.
- I filed survival/issue#270, which includes a figure that illustrates the issue, along with the minimal reproducible code for that figure.
Work-arounds
Before going on, I should mention some work-arounds for the issue I reported. Our goal was to create a data visualization that shows differences in prediction errors, between unsupervised penalty prediction using BIC, and supervised penalty prediction using survreg. Using either of the work-arounds discussed below, the “learned” regression line in the data visualization changes slightly, but it does not affect the overall message of the data visualization.
One work-around was changing survival::survreg (AFT loss) to
penaltyLearning::IntervalRegressionUnregularized (squared hinge
loss), resulting in a new data
viz.
Another work-around involves understanding how the survreg
optimization algorithm works. The survreg algorithm uses gradient
descent, which optimizes the coefficients to fit the train data
(maximize AFT log likelihood). Using the default scale=0 means to
also optimize the scale parameter. With the case of small data which are all
censored, there is an issue with optimizing the scale: the data may be separable, which
means there is a regression line that correctly predicts inside of
every interval label in the train set. In that case, decreasing the
scale parameter (subject to the constraint that it must stay positive)
will always increase the log likelihood. To work-around that
issue, you could use survreg(scale=1) which means to fix the scale
parameter to the (relatively large) value of 1, instead of optimizing
it in gradient descent.
Installing old survival
I seemed to remember that code working when I prepared my useR’17
presentation, so I suspected that a bug was introduced in survreg
some time after that. When exactly?
We can look at survival Archive to see when each version appeared on CRAN. And we can download those versions, and try to run the code.
First, we try to install an old version of survival, 2.40-1 from 2016-10-30:
(base) tdhock@tdhock-MacBook:~/R/survival[master]$ R -e 'u <- "https://cloud.r-project.org/src/contrib/Archive/survival/survival_2.40-1.tar.gz";download.file(u, l <- file.path("~/R",basename(u)));install.packages(l,repos=NULL)'
R version 4.4.1 (2024-06-14) -- "Race for Your Life"
Copyright (C) 2024 The R Foundation for Statistical Computing
Platform: x86_64-pc-linux-gnu
R est un logiciel libre livré sans AUCUNE GARANTIE.
Vous pouvez le redistribuer sous certaines conditions.
Tapez 'license()' ou 'licence()' pour plus de détails.
R est un projet collaboratif avec de nombreux contributeurs.
Tapez 'contributors()' pour plus d'information et
'citation()' pour la façon de le citer dans les publications.
Tapez 'demo()' pour des démonstrations, 'help()' pour l'aide
en ligne ou 'help.start()' pour obtenir l'aide au format HTML.
Tapez 'q()' pour quitter R.
Le chargement a nécessité le package : grDevices
> u <- "https://cloud.r-project.org/src/contrib/Archive/survival/survival_2.40-1.tar.gz";download.file(u, l <- file.path("~/R",basename(u)));install.packages(l,repos=NULL)
essai de l'URL 'https://cloud.r-project.org/src/contrib/Archive/survival/survival_2.40-1.tar.gz'
Content type 'application/x-gzip' length 4872012 bytes (4.6 MB)
==================================================
downloaded 4.6 MB
Le chargement a nécessité le package : grDevices
* installing *source* package ‘survival’ ...
** package ‘survival’ correctement décompressé et sommes MD5 vérifiées
** using staged installation
** libs
using C compiler: ‘gcc (GCC) 12.3.0’
gcc -I"/home/tdhock/lib/R/include" -DNDEBUG -I/usr/local/include -fpic -g -O2 -c agexact.c -o agexact.o
Dans le fichier inclus depuis agexact.c:53:
survproto.h:12:14: erreur: nom de type « Sint » inconnu; vouliez-vous utiliser « uint » ?
12 | void agfit5a(Sint *nusedx, Sint *nvarx, double *yy,
| ^~~~
| uint
...
agexact.c:59:14: erreur: nom de type « Sint » inconnu; vouliez-vous utiliser « uint » ?
59 | Sint *work2, double *eps, double *tol_chol, double *sctest)
| ^~~~
| uint
make: *** [/home/tdhock/lib/R/etc/Makeconf:195 : agexact.o] Erreur 1
ERROR: compilation failed for package ‘survival’
* removing ‘/home/tdhock/lib/R/library/survival’
* restoring previous ‘/home/tdhock/lib/R/library/survival’
Message d'avis :
Dans install.packages(l, repos = NULL) :
l'installation du package ‘/home/tdhock/R/survival_2.40-1.tar.gz’ a eu un statut de sortie non nul
>
>
We see from the output above that there are compilation errors using R-4.4.1 to try to install survival 2.40-1.
Below we try the same command, but with an older version of R.
(base) tdhock@tdhock-MacBook:~/R/survival[master]$ ~/R/R-3.6.3/bin/R -e 'u <- "https://cloud.r-project.org/src/contrib/Archive/survival/survival_2.40-1.tar.gz";download.file(u, l <- file.path("~/R",basename(u)));install.packages(l,repos=NULL)'
R version 3.6.3 (2020-02-29) -- "Holding the Windsock"
Copyright (C) 2020 The R Foundation for Statistical Computing
Platform: x86_64-pc-linux-gnu (64-bit)
R est un logiciel libre livré sans AUCUNE GARANTIE.
Vous pouvez le redistribuer sous certaines conditions.
Tapez 'license()' ou 'licence()' pour plus de détails.
R est un projet collaboratif avec de nombreux contributeurs.
Tapez 'contributors()' pour plus d'information et
'citation()' pour la façon de le citer dans les publications.
Tapez 'demo()' pour des démonstrations, 'help()' pour l'aide
en ligne ou 'help.start()' pour obtenir l'aide au format HTML.
Tapez 'q()' pour quitter R.
Le chargement a nécessité le package : grDevices
> u <- "https://cloud.r-project.org/src/contrib/Archive/survival/survival_2.40-1.tar.gz";download.file(u, l <- file.path("~/R",basename(u)));install.packages(l,repos=NULL)
essai de l'URL 'https://cloud.r-project.org/src/contrib/Archive/survival/survival_2.40-1.tar.gz'
Content type 'application/x-gzip' length 4872012 bytes (4.6 MB)
==================================================
downloaded 4.6 MB
Installation du package dans ‘/home/tdhock/R/x86_64-pc-linux-gnu-library/3.6’
(car ‘lib’ n'est pas spécifié)
Le chargement a nécessité le package : grDevices
* installing *source* package ‘survival’ ...
** package ‘survival’ correctement décompressé et sommes MD5 vérifiées
** using staged installation
** libs
gcc -I"/home/tdhock/R/R-3.6.3/include" -DNDEBUG -march=core2 -fpic -march=core2 -c agexact.c -o agexact.o
gcc -I"/home/tdhock/R/R-3.6.3/include" -DNDEBUG -march=core2 -fpic -march=core2 -c agfit4.c -o agfit4.o
gcc -I"/home/tdhock/R/R-3.6.3/include" -DNDEBUG -march=core2 -fpic -march=core2 -c agfit5.c -o agfit5.o
gcc -I"/home/tdhock/R/R-3.6.3/include" -DNDEBUG -march=core2 -fpic -march=core2 -c agmart.c -o agmart.o
gcc -I"/home/tdhock/R/R-3.6.3/include" -DNDEBUG -march=core2 -fpic -march=core2 -c agmart2.c -o agmart2.o
gcc -I"/home/tdhock/R/R-3.6.3/include" -DNDEBUG -march=core2 -fpic -march=core2 -c agmart3.c -o agmart3.o
gcc -I"/home/tdhock/R/R-3.6.3/include" -DNDEBUG -march=core2 -fpic -march=core2 -c agscore.c -o agscore.o
gcc -I"/home/tdhock/R/R-3.6.3/include" -DNDEBUG -march=core2 -fpic -march=core2 -c agsurv3.c -o agsurv3.o
gcc -I"/home/tdhock/R/R-3.6.3/include" -DNDEBUG -march=core2 -fpic -march=core2 -c agsurv4.c -o agsurv4.o
gcc -I"/home/tdhock/R/R-3.6.3/include" -DNDEBUG -march=core2 -fpic -march=core2 -c agsurv5.c -o agsurv5.o
gcc -I"/home/tdhock/R/R-3.6.3/include" -DNDEBUG -march=core2 -fpic -march=core2 -c chinv2.c -o chinv2.o
gcc -I"/home/tdhock/R/R-3.6.3/include" -DNDEBUG -march=core2 -fpic -march=core2 -c chinv3.c -o chinv3.o
gcc -I"/home/tdhock/R/R-3.6.3/include" -DNDEBUG -march=core2 -fpic -march=core2 -c cholesky2.c -o cholesky2.o
gcc -I"/home/tdhock/R/R-3.6.3/include" -DNDEBUG -march=core2 -fpic -march=core2 -c cholesky3.c -o cholesky3.o
gcc -I"/home/tdhock/R/R-3.6.3/include" -DNDEBUG -march=core2 -fpic -march=core2 -c chsolve2.c -o chsolve2.o
gcc -I"/home/tdhock/R/R-3.6.3/include" -DNDEBUG -march=core2 -fpic -march=core2 -c chsolve3.c -o chsolve3.o
gcc -I"/home/tdhock/R/R-3.6.3/include" -DNDEBUG -march=core2 -fpic -march=core2 -c concordance1.c -o concordance1.o
gcc -I"/home/tdhock/R/R-3.6.3/include" -DNDEBUG -march=core2 -fpic -march=core2 -c cox_Rcallback.c -o cox_Rcallback.o
gcc -I"/home/tdhock/R/R-3.6.3/include" -DNDEBUG -march=core2 -fpic -march=core2 -c coxcount1.c -o coxcount1.o
gcc -I"/home/tdhock/R/R-3.6.3/include" -DNDEBUG -march=core2 -fpic -march=core2 -c coxdetail.c -o coxdetail.o
gcc -I"/home/tdhock/R/R-3.6.3/include" -DNDEBUG -march=core2 -fpic -march=core2 -c coxexact.c -o coxexact.o
gcc -I"/home/tdhock/R/R-3.6.3/include" -DNDEBUG -march=core2 -fpic -march=core2 -c coxfit5.c -o coxfit5.o
gcc -I"/home/tdhock/R/R-3.6.3/include" -DNDEBUG -march=core2 -fpic -march=core2 -c coxfit6.c -o coxfit6.o
gcc -I"/home/tdhock/R/R-3.6.3/include" -DNDEBUG -march=core2 -fpic -march=core2 -c coxmart.c -o coxmart.o
gcc -I"/home/tdhock/R/R-3.6.3/include" -DNDEBUG -march=core2 -fpic -march=core2 -c coxmart2.c -o coxmart2.o
gcc -I"/home/tdhock/R/R-3.6.3/include" -DNDEBUG -march=core2 -fpic -march=core2 -c coxph_wtest.c -o coxph_wtest.o
gcc -I"/home/tdhock/R/R-3.6.3/include" -DNDEBUG -march=core2 -fpic -march=core2 -c coxsafe.c -o coxsafe.o
gcc -I"/home/tdhock/R/R-3.6.3/include" -DNDEBUG -march=core2 -fpic -march=core2 -c coxscho.c -o coxscho.o
gcc -I"/home/tdhock/R/R-3.6.3/include" -DNDEBUG -march=core2 -fpic -march=core2 -c coxscore.c -o coxscore.o
gcc -I"/home/tdhock/R/R-3.6.3/include" -DNDEBUG -march=core2 -fpic -march=core2 -c dmatrix.c -o dmatrix.o
gcc -I"/home/tdhock/R/R-3.6.3/include" -DNDEBUG -march=core2 -fpic -march=core2 -c doloop.c -o doloop.o
gcc -I"/home/tdhock/R/R-3.6.3/include" -DNDEBUG -march=core2 -fpic -march=core2 -c finegray.c -o finegray.o
gcc -I"/home/tdhock/R/R-3.6.3/include" -DNDEBUG -march=core2 -fpic -march=core2 -c init.c -o init.o
gcc -I"/home/tdhock/R/R-3.6.3/include" -DNDEBUG -march=core2 -fpic -march=core2 -c pyears1.c -o pyears1.o
gcc -I"/home/tdhock/R/R-3.6.3/include" -DNDEBUG -march=core2 -fpic -march=core2 -c pyears2.c -o pyears2.o
gcc -I"/home/tdhock/R/R-3.6.3/include" -DNDEBUG -march=core2 -fpic -march=core2 -c pyears3b.c -o pyears3b.o
gcc -I"/home/tdhock/R/R-3.6.3/include" -DNDEBUG -march=core2 -fpic -march=core2 -c pystep.c -o pystep.o
gcc -I"/home/tdhock/R/R-3.6.3/include" -DNDEBUG -march=core2 -fpic -march=core2 -c survConcordance.c -o survConcordance.o
gcc -I"/home/tdhock/R/R-3.6.3/include" -DNDEBUG -march=core2 -fpic -march=core2 -c survdiff2.c -o survdiff2.o
gcc -I"/home/tdhock/R/R-3.6.3/include" -DNDEBUG -march=core2 -fpic -march=core2 -c survfit4.c -o survfit4.o
gcc -I"/home/tdhock/R/R-3.6.3/include" -DNDEBUG -march=core2 -fpic -march=core2 -c survfitci.c -o survfitci.o
gcc -I"/home/tdhock/R/R-3.6.3/include" -DNDEBUG -march=core2 -fpic -march=core2 -c survpenal.c -o survpenal.o
gcc -I"/home/tdhock/R/R-3.6.3/include" -DNDEBUG -march=core2 -fpic -march=core2 -c survreg6.c -o survreg6.o
gcc -I"/home/tdhock/R/R-3.6.3/include" -DNDEBUG -march=core2 -fpic -march=core2 -c survreg7.c -o survreg7.o
gcc -I"/home/tdhock/R/R-3.6.3/include" -DNDEBUG -march=core2 -fpic -march=core2 -c survregc1.c -o survregc1.o
gcc -I"/home/tdhock/R/R-3.6.3/include" -DNDEBUG -march=core2 -fpic -march=core2 -c survregc2.c -o survregc2.o
gcc -I"/home/tdhock/R/R-3.6.3/include" -DNDEBUG -march=core2 -fpic -march=core2 -c survsplit.c -o survsplit.o
gcc -I"/home/tdhock/R/R-3.6.3/include" -DNDEBUG -march=core2 -fpic -march=core2 -c tmerge.c -o tmerge.o
gcc -shared -L/home/tdhock/R/R-3.6.3/lib -L/usr/local/lib -o survival.so agexact.o agfit4.o agfit5.o agmart.o agmart2.o agmart3.o agscore.o agsurv3.o agsurv4.o agsurv5.o chinv2.o chinv3.o cholesky2.o cholesky3.o chsolve2.o chsolve3.o concordance1.o cox_Rcallback.o coxcount1.o coxdetail.o coxexact.o coxfit5.o coxfit6.o coxmart.o coxmart2.o coxph_wtest.o coxsafe.o coxscho.o coxscore.o dmatrix.o doloop.o finegray.o init.o pyears1.o pyears2.o pyears3b.o pystep.o survConcordance.o survdiff2.o survfit4.o survfitci.o survpenal.o survreg6.o survreg7.o survregc1.o survregc2.o survsplit.o tmerge.o -L/home/tdhock/R/R-3.6.3/lib -lR
installing to /home/tdhock/R/x86_64-pc-linux-gnu-library/3.6/00LOCK-survival/00new/survival/libs
** R
** data
*** moving datasets to lazyload DB
** inst
** byte-compile and prepare package for lazy loading
Le chargement a nécessité le package : grDevices
** help
*** installing help indices
** building package indices
** installing vignettes
** testing if installed package can be loaded from temporary location
Le chargement a nécessité le package : grDevices
** checking absolute paths in shared objects and dynamic libraries
** testing if installed package can be loaded from final location
Le chargement a nécessité le package : grDevices
** testing if installed package keeps a record of temporary installation path
* DONE (survival)
Above we see that R-3.6.3 works for compiling the old version of survival.
Visualizing survival results on test data
Next, we need some R code to test whether or not the currently installed version of survival has the issue. We use the code below, which was adapted from the issue discussed earlier.
library(survival)
(full.df <- structure(list(id = c(1L, 1L, 1L, 1L, 1L, 1L, 4L, 4L, 4L, 4L,
4L, 4L, 6L, 6L, 6L, 6L, 6L, 6L, 8L, 8L, 8L, 8L, 8L, 8L, 10L,
10L, 10L, 10L, 10L, 10L, 11L, 11L, 11L, 11L, 11L, 11L, 13L, 13L,
13L, 13L, 13L, 13L, 120L, 120L, 120L, 120L, 120L, 120L), lo = c(0.195727051766757,
-1.1087553605429, -1.00065642410924, -0.995123241844028, -Inf,
-1.81065319423307, -Inf, -Inf, -Inf, -Inf, -Inf, -Inf, -0.694428755856871,
-2.4887060208859, -3.44756655828744, -3.03711252743929, -3.14523459850271,
-0.892319064459033, -0.552651313503626, -Inf, -2.43732789951484,
-1.24650632092422, -Inf, -Inf, -1.50791410241527, -2.24743277679978,
-2.37206425088304, -3.83417364430955, -2.60448041546997, -1.44599430482559,
-Inf, -0.456570392651195, -0.249593970642512, -Inf, 0.552800358347374,
1.03693235764513, 0.366866754402124, -2.52677122267308, -2.48507949525124,
-1.54283966348088, -1.59313273785538, -2.44940446306835, -0.513882229855861,
-3.450873922835, -3.02209394810166, -3.97458092881313, -3.57901135468839,
-2.1845818324618), hi = c(Inf, Inf, Inf, Inf, 2.06739808397873,
Inf, 3.26518538624204, 1.13643272732406, 2.27284346410842, 1.38565083489553,
0.492585941448176, 2.13713710441581, Inf, Inf, Inf, Inf, Inf,
Inf, Inf, 0.937658965079102, Inf, Inf, 3.17164978103177, 3.0679625505633,
Inf, Inf, Inf, Inf, Inf, Inf, 2.39473287695651, Inf, Inf, 0.730793396188341,
Inf, Inf, Inf, Inf, Inf, Inf, Inf, Inf, Inf, Inf, Inf, Inf, Inf,
Inf), feature = c(-2.65456939094638, -2.42126427679538, -2.0705540906376,
-2.41236133374335, -2.38707136376868, -2.50995205248869, -2.36922164775071,
-2.39136308372706, -2.35954108440544, -2.52855213967643, -2.61717785427761,
-2.32148054113571, -2.90042209374967, -2.81341071676004, -2.90042209374967,
-2.7646205525906, -2.97592964625781, -2.3434070875143, -2.87790396885268,
-2.92818004477678, -2.91781583734955, -3.04532040079498, -2.44619735876083,
-2.36685302923454, -3.17008566069877, -2.7646205525906, -2.54465665419377,
-3.12356564506388, -3.20645330486964, -2.39689577246529, -2.43836426623075,
-2.17187107950722, -2.52267471748792, -2.59336839058434, -1.84434871178509,
-1.9783687469636, -3.01613697015902, -3.0756635490299, -2.93034936935621,
-2.94970009845407, -2.89011456307748, -2.87110297166178, -3.01593498087151,
-3.01593498087151, -2.94694210938456, -2.71055333132033, -3.11226608980994,
-2.68824757380603)), class = "data.frame", row.names = c(NA,
-48L)))
## id lo hi feature
## 1 1 0.1957271 Inf -2.654569
## 2 1 -1.1087554 Inf -2.421264
## 3 1 -1.0006564 Inf -2.070554
## 4 1 -0.9951232 Inf -2.412361
## 5 1 -Inf 2.0673981 -2.387071
## 6 1 -1.8106532 Inf -2.509952
## 7 4 -Inf 3.2651854 -2.369222
## 8 4 -Inf 1.1364327 -2.391363
## 9 4 -Inf 2.2728435 -2.359541
## 10 4 -Inf 1.3856508 -2.528552
## 11 4 -Inf 0.4925859 -2.617178
## 12 4 -Inf 2.1371371 -2.321481
## 13 6 -0.6944288 Inf -2.900422
## 14 6 -2.4887060 Inf -2.813411
## 15 6 -3.4475666 Inf -2.900422
## 16 6 -3.0371125 Inf -2.764621
## 17 6 -3.1452346 Inf -2.975930
## 18 6 -0.8923191 Inf -2.343407
## 19 8 -0.5526513 Inf -2.877904
## 20 8 -Inf 0.9376590 -2.928180
## 21 8 -2.4373279 Inf -2.917816
## 22 8 -1.2465063 Inf -3.045320
## 23 8 -Inf 3.1716498 -2.446197
## 24 8 -Inf 3.0679626 -2.366853
## 25 10 -1.5079141 Inf -3.170086
## 26 10 -2.2474328 Inf -2.764621
## 27 10 -2.3720643 Inf -2.544657
## 28 10 -3.8341736 Inf -3.123566
## 29 10 -2.6044804 Inf -3.206453
## 30 10 -1.4459943 Inf -2.396896
## 31 11 -Inf 2.3947329 -2.438364
## 32 11 -0.4565704 Inf -2.171871
## 33 11 -0.2495940 Inf -2.522675
## 34 11 -Inf 0.7307934 -2.593368
## 35 11 0.5528004 Inf -1.844349
## 36 11 1.0369324 Inf -1.978369
## 37 13 0.3668668 Inf -3.016137
## 38 13 -2.5267712 Inf -3.075664
## 39 13 -2.4850795 Inf -2.930349
## 40 13 -1.5428397 Inf -2.949700
## 41 13 -1.5931327 Inf -2.890115
## 42 13 -2.4494045 Inf -2.871103
## 43 120 -0.5138822 Inf -3.015935
## 44 120 -3.4508739 Inf -3.015935
## 45 120 -3.0220939 Inf -2.946942
## 46 120 -3.9745809 Inf -2.710553
## 47 120 -3.5790114 Inf -3.112266
## 48 120 -2.1845818 Inf -2.688248
write.csv(full.df, "~/R/survreg-data.csv", row.names=FALSE)
The code above defines the data set which is known to cause the issue, and saves it to a CSV file on disk. The data frame has one row for every labeled change-point problem, and four columns:
idis an identifier for where the data set came from.loandhiare the lower/upper limits of good log(penalty) values for this particular labeled change-point problem.featureis an input/covariate that should be used in a linear model to predict a log(penalty) value, which ideally falls betweenloandhifor every labeled change-point problem.
The code below defines two different functions for fitting linear regression models to the train data with censored outputs.
model.fun.list <- list(
survreg=function(train.df, sc){
fit <- with(train.df, survreg(
Surv(lo, hi, type="interval2") ~ feature,
scale=sc,
dist="gaussian"))
w <- coef(fit)
w["scale.out"] <- fit$scale
w
},
penaltyLearning=function(train.df, sc){
fit <- with(train.df, penaltyLearning::IntervalRegressionUnregularized(
cbind(feature), cbind(lo,hi), margin=sc))
w <- coef(fit)[,"0"]
w["scale.out"] <- sc
w
})
The functions above compute the learned weights, using either
survreg (AFT loss), or IntervalRegressionUnregularized (squared
hinge loss). The code below uses these learning algorithms on three
different data sets (defined by for loop over extra.id), and three
different models (defined by for loop over model.i).
model.df <- rbind(
data.frame(pkg="penaltyLearning", scale=1),
data.frame(pkg="survreg", scale=0),
data.frame(pkg="survreg", scale=1))
common.ids <- c(1, 4, 6, 8, 10, 11)
dot.df.list <- list()
coef.df.list <- list()
for(extra.id in c(0,13,120)){
sub.df <- transform(
subset(full.df, id %in% c(common.ids, extra.id)),
type=ifelse(id %in% common.ids, "common", "extra"))
X <- with(sub.df, cbind(1, feature))
print(chol(t(X) %*% X))
for(model.i in 1:nrow(model.df)){
model.row <- model.df[model.i,]
rownames(model.row) <- NULL
model.fun <- model.fun.list[[model.row$pkg]]
this.df <- data.frame(extra.id, model.row, warn.text="")
set.warn <- function(w)this.df$warn.text <<- w$message
set.coefs <- function(){
weight.vec <- model.fun(sub.df, model.row$scale)
this.df$intercept <<- weight.vec[["(Intercept)"]]
this.df$slope <<- weight.vec[["feature"]]
this.df$scale.out <<- weight.vec[["scale.out"]]
}
withCallingHandlers(set.coefs(), warning=set.warn)
coef.df.list[[paste(extra.id, model.i)]] <- this.df
dot.df.list[[paste(extra.id, model.i)]] <- data.frame(
extra.id, model.row,
with(sub.df, data.frame(feature, type, rbind(
data.frame(limit="lo", output=lo),
data.frame(limit="hi", output=hi))))
)
}
}
## feature
## 6 -15.513144
## feature 0 1.966648
## Warning in survreg.fit(X, Y, weights, offset, init = init, controlvals = control, : Ran out of iterations and did not
## converge
## feature
## 6.480741 -17.098652
## feature 0.000000 2.145202
## feature
## 6.480741 -17.061128
## feature 0.000000 2.139963
## Warning in survreg.fit(X, Y, weights, offset, init = init, controlvals = control, : Ran out of iterations and did not
## converge
dot.df <- subset(do.call(rbind, dot.df.list), is.finite(output))
(coef.df <- do.call(rbind, coef.df.list))
## extra.id pkg scale warn.text intercept slope scale.out
## 0 1 0 penaltyLearning 1 4.509705 1.5654255 1.000000000
## 0 2 0 survreg 0 Ran out of iterations and did not converge NA NA 0.002830596
## 0 3 0 survreg 1 4.163601 1.3984400 1.000000000
## 13 1 13 penaltyLearning 1 3.287217 1.0413089 1.000000000
## 13 2 13 survreg 0 2.580365 0.7560768 0.227619730
## 13 3 13 survreg 1 2.908041 0.8542918 1.000000000
## 120 1 120 penaltyLearning 1 3.914680 1.3137750 1.000000000
## 120 2 120 survreg 0 Ran out of iterations and did not converge 5.990912 2.1580340 0.041260001
## 120 3 120 survreg 1 3.583528 1.1475425 1.000000000
The result above is a table with one row per model and data set, with columns
extra.ididentifies the data set,pkgis the learning algorithm,scale=0means optimized,scale=1means fixed,warn.textis the warning that resulted,interceptandslopeare the learned linear model coefficients.
It is clear from the table above that finite coefficients are often
returned without warning, but survreg with scale=0 can run out of
iterations, and return missing/NA coefficients. Below, we visualize
these results along with the training data.
library(ggplot2)
ggplot()+
theme_bw()+
geom_abline(aes(
slope=slope, intercept=intercept),
data=coef.df)+
facet_grid(pkg + scale ~ extra.id, labeller=label_both)+
geom_point(aes(
feature, output, fill=limit, color=type),
shape=21,
data=dot.df)+
scale_color_manual(values=c(
common="white",
extra="black"))+
geom_text(aes(
-3.2, 5.5, label=sprintf(
"slope=%.2f intercept=%.2f scale=%.4f\n%s", slope, intercept, scale.out, warn.text)),
vjust=1,
hjust=0,
size=3,
data=coef.df)
## Warning: Removed 1 row containing missing values or values outside the scale range (`geom_abline()`).
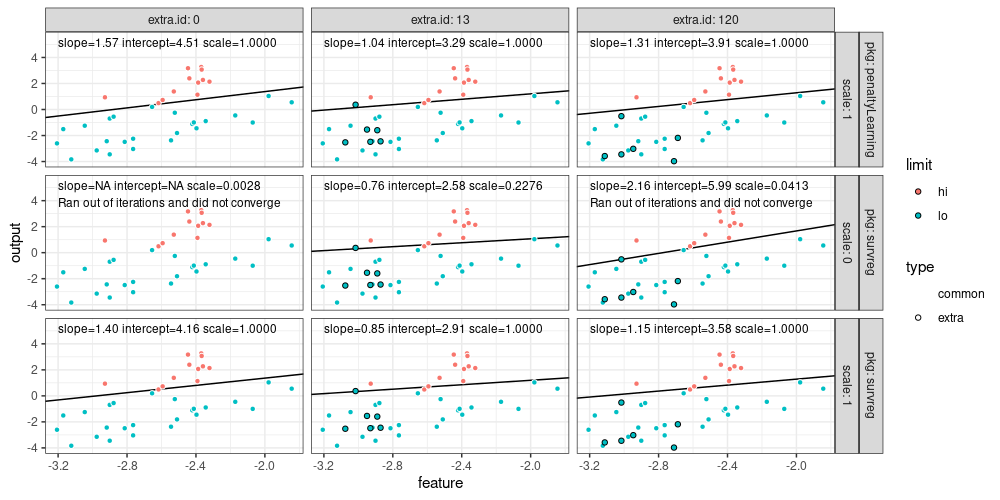
The figure above shows that data are separable when there are convergence warnings.
Testing survival
The results in the figure above can be summarized using the code
below, which only runs survreg with the default scale, meaning scale
parameter optimized in gradient descent.
library(survival)
full.df <- read.csv("~/R/survreg-data.csv")
coef.df.list <- list()
for(extra.id in c(0,13,120)){
sub.df <- subset(full.df, id %in% c(1, 4, 6, 8, 10, 11, extra.id))
this.df <- data.frame(
R=R.version$version.string,
survival=packageVersion("survival"), extra.id, warn.text="")
set.warn <- function(w)this.df$warn.text <<- w$message
set.coefs <- function(){
weight.vec <- coef(with(sub.df, survreg(
Surv(lo, hi, type="interval2") ~ feature,
dist="gaussian")))
this.df$intercept <<- weight.vec[["(Intercept)"]]
this.df$slope <<- weight.vec[["feature"]]
}
try.out <- withCallingHandlers(set.coefs(), warning=set.warn)
coef.df.list[[paste(extra.id)]] <- this.df
}
## Warning in survreg.fit(X, Y, weights, offset, init = init, controlvals = control, : Ran out of iterations and did not
## converge
## Warning in survreg.fit(X, Y, weights, offset, init = init, controlvals = control, : Ran out of iterations and did not
## converge
(coef.df <- do.call(rbind, coef.df.list))
## R survival extra.id warn.text intercept slope
## 0 R version 4.4.1 (2024-06-14) 3.7.1 0 Ran out of iterations and did not converge NA NA
## 13 R version 4.4.1 (2024-06-14) 3.7.1 13 2.580365 0.7560768
## 120 R version 4.4.1 (2024-06-14) 3.7.1 120 Ran out of iterations and did not converge 5.990912 2.1580340
out.csv <- gsub("[ ()]", "_", paste0(
"~/R/survreg-data/",
R.version$version.string,
"survival",
packageVersion("survival"),
".csv"))
dir.create(dirname(out.csv), showWarnings = FALSE)
write.csv(coef.df, out.csv, row.names=FALSE)
The code above stores the versions of R and survival, along with the result of running survreg on the test data.
I copied the code block above to ~/R/survreg-data.R, and
I ran it using the old version of R that I have on my system:
(base) tdhock@tdhock-MacBook:~/R$ ~/R/R-3.6.3/bin/R --vanilla < ~/R/survreg-data.R
R version 3.6.3 (2020-02-29) -- "Holding the Windsock"
Copyright (C) 2020 The R Foundation for Statistical Computing
Platform: x86_64-pc-linux-gnu (64-bit)
R est un logiciel libre livré sans AUCUNE GARANTIE.
Vous pouvez le redistribuer sous certaines conditions.
Tapez 'license()' ou 'licence()' pour plus de détails.
R est un projet collaboratif avec de nombreux contributeurs.
Tapez 'contributors()' pour plus d'information et
'citation()' pour la façon de le citer dans les publications.
Tapez 'demo()' pour des démonstrations, 'help()' pour l'aide
en ligne ou 'help.start()' pour obtenir l'aide au format HTML.
Tapez 'q()' pour quitter R.
> library(survival)
> full.df <- read.csv("~/R/survreg-data.csv")
> coef.df.list <- list()
> for(extra.id in c(0,13,120)){
+ sub.df <- subset(full.df, id %in% c(1, 4, 6, 8, 10, 11, extra.id))
+ this.df <- data.frame(
+ R=R.version$version.string,
+ survival=packageVersion("survival"), extra.id, warn.text="")
+ set.warn <- function(w)this.df$warn.text <<- w$message
+ set.coefs <- function(){
+ weight.vec <- coef(with(sub.df, survreg(
+ Surv(lo, hi, type="interval2") ~ feature,
+ dist="gaussian")))
+ this.df$intercept <<- weight.vec[["(Intercept)"]]
+ this.df$slope <<- weight.vec[["feature"]]
+ }
+ try.out <- withCallingHandlers(set.coefs(), warning=set.warn)
+ coef.df.list[[paste(extra.id)]] <- this.df
+ }
Warning messages:
1: In survreg.fit(X, Y, weights, offset, init = init, controlvals = control, :
Ran out of iterations and did not converge
2: In survreg.fit(X, Y, weights, offset, init = init, controlvals = control, :
Ran out of iterations and did not converge
> (coef.df <- do.call(rbind, coef.df.list))
R survival extra.id
0 R version 3.6.3 (2020-02-29) 3.1.8 0
13 R version 3.6.3 (2020-02-29) 3.1.8 13
120 R version 3.6.3 (2020-02-29) 3.1.8 120
warn.text intercept slope
0 Ran out of iterations and did not converge 6.916020 2.4876223
13 2.580365 0.7560768
120 Ran out of iterations and did not converge 5.990912 2.1580340
> out.csv <- gsub("[ ()]", "_", paste0(
+ "~/R/survreg-data/",
+ R.version$version.string,
+ "survival",
+ packageVersion("survival"),
+ ".csv"))
> dir.create(dirname(out.csv))
Warning message:
In dir.create(dirname(out.csv)) : '/home/tdhock/R/survreg-data' existe déjà
> write.csv(coef.df, out.csv, row.names=FALSE)
>
The output above shows that all coefficients are finite, so the problem does not exist in this old version. Same for 3.5-0. But there are missing coefs in 3.6-4. So the change must have started somewhere in between. Here are the results I computed:
result.df.list <- list()
for(f.csv in Sys.glob("../assets/survreg-data/*.csv")){
result.df.list[[f.csv]] <- read.csv(f.csv)
}
result.df <- do.call(rbind, result.df.list)
rownames(result.df) <- NULL
subset(result.df, extra.id==0)
## R survival extra.id warn.text intercept slope
## 1 R version 3.6.3 (2020-02-29) 3.1.8 0 Ran out of iterations and did not converge 6.91602 2.487622
## 4 R version 3.6.3 (2020-02-29) 3.3.0 0 Ran out of iterations and did not converge 6.91602 2.487622
## 7 R version 3.6.3 (2020-02-29) 3.5.0 0 Ran out of iterations and did not converge 6.91602 2.487622
## 10 R version 3.6.3 (2020-02-29) 3.5.3 0 Ran out of iterations and did not converge NA NA
## 13 R version 3.6.3 (2020-02-29) 3.5.5 0 Ran out of iterations and did not converge NA NA
## 16 R version 3.6.3 (2020-02-29) 3.5.7 0 Ran out of iterations and did not converge NA NA
## 19 R version 3.6.3 (2020-02-29) 3.6.4 0 Ran out of iterations and did not converge NA NA
## 22 R version 4.4.1 (2024-06-14) 3.6.4 0 Ran out of iterations and did not converge NA NA
It can be seen above that the missing values started happening some
time between survival 3.5.0 and 3.5.3.
git bisect
In the last section, we identified which CRAN release version of survival (3.5.3) was responsible for introducing the missing values bug. We started with the most recent version, which had the missing values, and then we went back in time until we found a version which did not have the bug (3.1.8). Then we tested intermediate versions until we found adjacent versions, one with missing, other without.
The log time algorithm to find these adjacent versions is called “bisection.” The idea is to always test the version in the middle of the versions that you already know (and it works because of the continuity between versions – there is one change that introduced the bug). In our case there were 20 versions, so we test the one in the middle, now there are only 10, next step there are only 5, then 3 (or 2), then 2 (or 1), then finally only one left to test. So we test 7 versions using this algorithm (asymptotically log2 of the number of versions in between known good/old and bad/new versions), which is much faster than 20 versions which we would need to test using linear search.
Now that we have found the CRAN release version, we may ask, what git
commit is responsible? We can use the git bisect command, as I did
to implement the data.table rev-dep
checker. That
script returns 0 if it works, 1 if it fails. The analogous script for
this blog is below, which has code for testing whether or not we get a
missing value:
install.packages(".",repos=NULL)
library(survival)
full.df <- read.csv("~/R/survreg-data.csv")
sub.df <- subset(full.df, id %in% c(1, 4, 6, 8, 10, 11, 0))
weight.vec <- coef(with(sub.df, survreg(
Surv(lo, hi, type="interval2") ~ feature,
dist="gaussian")))
q(status=if(is.na(weight.vec[["feature"]]))1 else 0)
I saved the code block above to the file ~/R/survreg-test.R so we can use it in the context of git bisect. Next step is to find git commits corresponding to the old/new versions of interest.
The survival Archive page on
CRAN says
that the old/new versions of interest were released on 2023-01-09 23:50 and
2023-02-12 22:30. The code below computes the commits corresponding to these dates.
(base) tdhock@tdhock-MacBook:~/R/survival$ git checkout `git rev-list -1 --before="2023-01-09 23:50:00" master`
HEAD est maintenant sur dbf56fa Final updates for CRAN release
(base) tdhock@tdhock-MacBook:~/R/survival$ git checkout `git rev-list -1 --before="2023-02-12 22:30:00" master`
La position précédente de HEAD était sur dbf56fa Final updates for CRAN release
HEAD est maintenant sur 3f8f3e9 Add trust regions to coxph.
The output above shows the commits we can use, old=dbf56fa, new=3f8f3e9. The code below computes the bisect.
(base) tdhock@tdhock-MacBook:~/R/survival$ git bisect start
La position précédente de HEAD était sur 3f8f3e9 Add trust regions to coxph.
HEAD est maintenant sur dbf56fa Final updates for CRAN release
(base) tdhock@tdhock-MacBook:~/R/survival$ git bisect new 3f8f3e9
(base) tdhock@tdhock-MacBook:~/R/survival$ git bisect old dbf56fa
Bissection : 5 révisions à tester après ceci (à peu près 3 étapes)
[5c0d5d897c8747bb4117542e381490c2a0cb2172] Updates to the trust region discussion.
Lancement de 'Rscript' '/home/tdhock/R/survreg-test.R'
...
Dans survreg.fit(X, Y, weights, offset, init = init, controlvals = control, :
Ran out of iterations and did not converge
Bissection : 2 révisions à tester après ceci (à peu près 2 étapes)
[ede711c5fa4ce819801d856582369ee5eea94fd6] Minor changes to 3 routines, bug in fastkm.c
Lancement de 'Rscript' '/home/tdhock/R/survreg-test.R'
...
Dans survreg.fit(X, Y, weights, offset, init = init, controlvals = control, :
Ran out of iterations and did not converge
Bissection : 0 révision à tester après ceci (à peu près 1 étape)
[d373659174a1b843013c1f7ab750e5d876ccfd32] Merge
Lancement de 'Rscript' '/home/tdhock/R/survreg-test.R'
...
Dans survreg.fit(X, Y, weights, offset, init = init, controlvals = control, :
Ran out of iterations and did not converge
ede711c5fa4ce819801d856582369ee5eea94fd6 is the first new commit
commit ede711c5fa4ce819801d856582369ee5eea94fd6
Author: Terry Therneau <terry.therneau@gmail.com>
Date: Fri Jan 20 17:03:33 2023 -0600
Minor changes to 3 routines, bug in fastkm.c
DESCRIPTION | 4 ++--
R/summary.coxph.R | 8 +++++---
R/survreg.R | 4 ++--
inst/NEWS.Rd | 7 +++++++
src/concordance5.c | 2 +-
src/fastkm.c | 10 +++++-----
6 files changed, 22 insertions(+), 13 deletions(-)
The output above shows the first commit that git found with the issue.
It has modifications to R/survreg.R, as shown below:
(base) tdhock@tdhock-MacBook:~/R/survival[master]$ git show ede711c5fa4ce819801d856582369ee5eea94fd6 R/survreg.R
commit ede711c5fa4ce819801d856582369ee5eea94fd6 (refs/bisect/new)
Author: Terry Therneau <terry.therneau@gmail.com>
Date: Fri Jan 20 17:03:33 2023 -0600
Minor changes to 3 routines, bug in fastkm.c
diff --git a/R/survreg.R b/R/survreg.R
index d4da378..edc9ad5 100644
--- a/R/survreg.R
+++ b/R/survreg.R
@@ -266,8 +266,8 @@ survreg <- function(formula, data, weights, subset, na.action,
# set singular coefficients to NA
# this is purposely not done until the residuals, etc. are computed
- singular <- (diag(fit$var)==0)[1:length(fit$coef)]
- if (any(singular)) fit$coeffients[singular] <- NA
+ singular <- (diag(fit$var)==0)[1:length(fit$coefficients)]
+ if (any(singular)) fit$coefficients[singular] <- NA
na.action <- attr(m, "na.action")
if (length(na.action)) fit$na.action <- na.action
The code above shows that a typo coeffients was changed to
coefficients, in a line that assigns NA, which explains why we see
missing values in survreg output (when singular). Does singular mean the same as separable? This may actually be
interpreted as a feature/fix, rather than a bug, because the scale of the solution
is undefined when the data are separable (several different scale parameters have the same max likelihood).
Conclusion
We have discussed the history of supervised change-point detection,
and its relationship with survival data analysis (censored
regression). We showed how to use survreg in R, and used git
bisect to find what version started to output missing values for
coefficients. A remaining question is for the extra.id=120 data set,
for which we run out of iterations (no convergence), yet we get finite
coefficients, and the scatterplot appears to be separable. It seems to
me that this is inconsistent with the results we observe for
extra.id=0 (no convergence, missing coefficients). I would suggest
that survreg should return missing values for both cases, or finite
values for both cases. More generally, if the data are separable, then
it is reasonable for survreg to return missing coefficients. On the
other hand, a warning is sufficient to alert the user, and returning
finite coefficients (last iteration of gradient descent before giving
up) still may be useful to the user.