Generate publications page
I have been updating my publications page by editing markdown for several years. Today I updated my self-citation database, TDH-refs.bib, to be consistent with that publications page. In this post we explore the extent to which it would be possible to generate the publications page, using the bib file as a source.
Parse bib into R
Parsing bibtex files is easy using regex. In fact, that is one of the
examples on ?nc::capture_all_str:
refs.bib <- "~/tdhock.github.io/assets/TDH-refs.bib"
refs.vec <- readLines(refs.bib)
at.lines <- grep("^@", refs.vec, value=TRUE)
str(at.lines)
## chr [1:73] "@unpublished{Agyapong2026poisson," "@unpublished{Agyapong2026fused," ...
The output above shows that there are currently 73 lines that start
with @ in the bib file. Below we use a regex to convert each item
into one row of a data table:
refs.dt <- nc::capture_all_str(
refs.vec,
"@",
type="[^{]+", tolower,
"[{]",
ref="[^,]+",
",\n",
fields="(?:.*\n)+?.*",
"[}]\\s*(?:$|\n)")
str(refs.dt)
## Classes 'data.table' and 'data.frame': 73 obs. of 3 variables:
## $ type : chr "unpublished" "unpublished" "unpublished" "unpublished" ...
## $ ref : chr "Agyapong2026poisson" "Agyapong2026fused" "Amoakohene2025asymptotic" "Oliveira2025governance" ...
## $ fields: chr " title={Identifying regimes where graphical models out-perform linear models for microbiome counts},\n note={"| __truncated__ " title={Fused Lasso Improves Accuracy of Co-occurrence Network Inference in Grouped Samples},\n note={Preprin"| __truncated__ " title={Asymptotic benchmarking using the atime package},\n author={Amoakohene, D and Chetia, A and Steinmach"| __truncated__ " title=,\n author={Oliveira, P a"| __truncated__ ...
## - attr(*, ".internal.selfref")=<externalptr>
The output above shows that the bib file was converted to a table with 73 rows.
Parsing fields
First we look at the number of lines with an equals sign, each of which is probably a field.
eq.lines <- grep("=", refs.vec, value=TRUE)
str(eq.lines)
## chr [1:561] " title={Identifying regimes where graphical models out-perform linear models for microbiome counts}," ...
Above we see 561 fields.
Below we parse the fields column:
strip <- function(x)gsub("^\\s*|,\\s*$", "", gsub('[{}"]', "", x))
field.pattern <- list(
"\\s*",
variable="[^= ]+", tolower,
"\\s*=",
value=".*", strip)
(refs.fields <- refs.dt[, nc::capture_all_str(
fields, field.pattern),
by=.(type, ref)])
## type ref variable
## <char> <char> <char>
## 1: unpublished Agyapong2026poisson title
## 2: unpublished Agyapong2026poisson note
## 3: unpublished Agyapong2026poisson author
## 4: unpublished Agyapong2026poisson links
## 5: unpublished Agyapong2026poisson year
## ---
## 557: article Doyon2008heritable number
## 558: article Doyon2008heritable pages
## 559: article Doyon2008heritable year
## 560: article Doyon2008heritable links
## 561: article Doyon2008heritable publisher
## value
## <char>
## 1: Identifying regimes where graphical models out-perform linear models for microbiome counts
## 2: In Progress
## 3: Daniel Agyapong and Julien Chiquet and Jane Marks and Toby Dylan Hocking
## 4: [Reproducible](https://github.com/EngineerDanny/pln_eval)
## 5: 2026
## ---
## 557: 6
## 558: 702--708
## 559: 2008
## 560: [Pubmed](http://www.ncbi.nlm.nih.gov/pubmed/18500334)
## 561: Nature Publishing Group US New York
Above we see 561 fields, consistent with the simpler grep parsing above.
If it is not consistent, we can use the code below to find out where:
(eq.dt <- nc::capture_first_vec(eq.lines, field.pattern))
## variable value
## <char> <char>
## 1: title Identifying regimes where graphical models out-perform linear models for microbiome counts
## 2: note In Progress
## 3: author Daniel Agyapong and Julien Chiquet and Jane Marks and Toby Dylan Hocking
## 4: links [Reproducible](https://github.com/EngineerDanny/pln_eval)
## 5: year 2026
## ---
## 557: number 6
## 558: pages 702--708
## 559: year 2008
## 560: links [Pubmed](http://www.ncbi.nlm.nih.gov/pubmed/18500334)
## 561: publisher Nature Publishing Group US New York
eq.dt[!refs.fields, on=.(variable,value)]
## Empty data.table (0 rows and 2 cols): variable,value
eq.counts <- eq.dt[, .(eq.count=.N), by=.(variable,value)]
refs.fields[, .(ref.count=.N), by=.(variable,value)][eq.counts,on=.(variable,value)][eq.count!=ref.count]
## Empty data.table (0 rows and 4 cols): variable,value,ref.count,eq.count
Verify clean
Normally there should not be any quotes or curly braces in fields:
cat(grep('[{}"]', refs.fields$value, value=TRUE), sep="\n\n")
Formatting the parsed data
The publications page is organized as follows
- chronologically, newest on top.
- heading
###for in progress, and each year. - bullet
-for each publication.
Each publication has
- Names like Last1 F1, Last2 F2.
- Title then period.
- Venue/publisher.
- then links.
The links at the end are not stored in the bib file, so that is not possible to output. (exercise for reader!)
But we can get the other info.
Venues
library(data.table)
## data.table 1.17.99 IN DEVELOPMENT built 2025-09-05 15:39:30 UTC using 3 threads (see ?getDTthreads). Latest news: r-datatable.com
## **********
## This development version of data.table was built more than 4 weeks ago. Please update: data.table::update_dev_pkg()
## **********
refs.wide <- dcast(refs.fields, type + ref ~ variable)
fwrite(refs.wide,"../assets/TDH-refs.csv")
refs.wide[, .(
type,
year,
journal=substr(journal,1,10),
vol=volume, num=number,
booktitle=substr(booktitle,1,10),
note=substr(note,1,10),
school=substr(school,1,10))]
## Key: <type>
## type year journal vol num booktitle note school
## <char> <char> <char> <char> <char> <char> <char> <char>
## 1: article 2021 Functional 35 4 <NA> <NA> <NA>
## 2: article 2025 BMC Bioinf 26 74 <NA> <NA> <NA>
## 3: article 2018 The Americ 103 4 <NA> <NA> <NA>
## 4: article 2022 Journal of 31 4 <NA> <NA> <NA>
## 5: article 2024 Journal of 1 3 <NA> e2024JH000 <NA>
## 6: article 2022 ACM Trans. 29 2 <NA> <NA> <NA>
## 7: article 2016 Clinical C 22 22 <NA> <NA> <NA>
## 8: article 2018 JNCI: Jour 110 10 <NA> <NA> <NA>
## 9: article 2018 Scientific 5 1 <NA> <NA> <NA>
## 10: article 2008 Nature bio 26 6 <NA> <NA> <NA>
## 11: article 2021 Computers 130 <NA> <NA> <NA> <NA>
## 12: article 2010 PLoS one 5 8 <NA> <NA> <NA>
## 13: article 2024 Environmen 19 11 <NA> <NA> <NA>
## 14: article 2023 IEEE Robot 8 8 <NA> <NA> <NA>
## 15: article 2023 Journal of 24 70 <NA> <NA> <NA>
## 16: article 2013 BMC Bioinf 14 164 <NA> <NA> <NA>
## 17: article 2013 Journal of 54 <NA> <NA> <NA> <NA>
## 18: article 2014 Bioinforma 30 11 <NA> <NA> <NA>
## 19: article 2017 Bioinforma 33 4 <NA> <NA> <NA>
## 20: article 2019 The R Jour 11 2 <NA> <NA> <NA>
## 21: article 2020 Journal of 21 87 <NA> <NA> <NA>
## 22: article 2021 The R Jour 13 1 <NA> <NA> <NA>
## 23: article 2022 Journal of 101 10 <NA> <NA> <NA>
## 24: article 2023 Computatio 38 <NA> <NA> <NA> <NA>
## 25: article 2019 Biostatist 21 4 <NA> <NA> <NA>
## 26: article 2024 Journal of 33 4 <NA> <NA> <NA>
## 27: article 2021 BMC Bioinf 22 323 <NA> <NA> <NA>
## 28: article 2017 Statistics 27 <NA> <NA> <NA> <NA>
## 29: article 2022 Biology Me 7 1 <NA> <NA> <NA>
## 30: article 2025 Statistics 35 <NA> <NA> <NA> <NA>
## 31: article 2023 Journal of 106 6 <NA> <NA> <NA>
## 32: article 2016 Leukemia 30 7 <NA> <NA> <NA>
## 33: article 2019 Journal of 28 2 <NA> <NA> <NA>
## 34: article 2014 Cancer Sci 105 7 <NA> <NA> <NA>
## 35: article 2023 Nature 618 <NA> <NA> DOI:10.103 <NA>
## 36: article 2024 Nature 627 8002 <NA> <NA> <NA>
## 37: article 2022 Journal of 31 2 <NA> <NA> <NA>
## 38: incollection 2022 <NA> <NA> <NA> Land Carbo <NA> <NA>
## 39: inproceedings 2022 <NA> <NA> <NA> 2022 Fourt <NA> <NA>
## 40: inproceedings 2022 <NA> <NA> <NA> 2022 fourt <NA> <NA>
## 41: inproceedings 2017 <NA> <NA> <NA> Advances i <NA> <NA>
## 42: inproceedings 2020 <NA> <NA> <NA> 2020 54th <NA> <NA>
## 43: inproceedings 2020 <NA> <NA> <NA> 2020 42nd <NA> <NA>
## 44: inproceedings 2011 <NA> <NA> <NA> 28th inter <NA> <NA>
## 45: inproceedings 2013 <NA> <NA> <NA> Proc. 30th <NA> <NA>
## 46: inproceedings 2015 <NA> <NA> <NA> Proc. 32nd <NA> <NA>
## 47: inproceedings 2020 <NA> 25 <NA> Proc. Paci <NA> <NA>
## 48: inproceedings 2022 <NA> <NA> <NA> 2022 Fourt <NA> <NA>
## 49: inproceedings 2021 <NA> <NA> <NA> 2021 IEEE <NA> <NA>
## 50: inproceedings 2023 <NA> <NA> <NA> 2023 Inter <NA> <NA>
## 51: phdthesis 2012 <NA> <NA> <NA> <NA> <NA> Ecole norm
## 52: unpublished 2025 <NA> <NA> <NA> <NA> Preprint a <NA>
## 53: unpublished 2026 <NA> <NA> <NA> <NA> In Progres <NA>
## 54: unpublished 2025 <NA> <NA> <NA> <NA> Under revi <NA>
## 55: unpublished 2024 <NA> <NA> <NA> <NA> Preprint a <NA>
## 56: unpublished 2015 <NA> <NA> <NA> <NA> Preprint a <NA>
## 57: unpublished 2016 <NA> <NA> <NA> <NA> Tutorial a <NA>
## 58: unpublished 2017 <NA> <NA> <NA> <NA> Tutorial a <NA>
## 59: unpublished 2023 <NA> <NA> <NA> <NA> In progres <NA>
## 60: unpublished 2025 <NA> <NA> <NA> <NA> Under revi <NA>
## 61: unpublished 2024 <NA> <NA> <NA> <NA> Preprint a <NA>
## 62: unpublished 2024 <NA> <NA> <NA> <NA> In progres <NA>
## 63: unpublished 2024 <NA> <NA> <NA> <NA> In progres <NA>
## 64: unpublished 2024 <NA> <NA> <NA> <NA> Preprint a <NA>
## 65: unpublished 2026 <NA> <NA> <NA> <NA> Abstract s <NA>
## 66: unpublished 2025 <NA> <NA> <NA> <NA> Preprint a <NA>
## 67: unpublished 2025 <NA> <NA> <NA> <NA> Preprint a <NA>
## 68: unpublished 2025 <NA> <NA> <NA> <NA> ICSME (Int <NA>
## 69: unpublished 2023 <NA> <NA> <NA> <NA> Preprint a <NA>
## 70: unpublished 2025 <NA> <NA> <NA> <NA> Under revi <NA>
## 71: unpublished 2024 <NA> <NA> <NA> <NA> Under revi <NA>
## 72: unpublished 2024 <NA> <NA> <NA> <NA> In progres <NA>
## 73: unpublished 2014 <NA> <NA> <NA> <NA> Preprint a <NA>
## type year journal vol num booktitle note school
## <char> <char> <char> <char> <char> <char> <char> <char>
As can be seen in the table above, we can use various fields to define the venue of publication:
article: most journal articles have avolumeandnumber. The only exception is a recently published article that has not yet been assigned a volume.incollectionandinproceedings: can usebooktitle.unpublished: can usenote.phdthesis: can useschool.
These rules are encoded below,
refs.wide[, venue := fcase(
type=="article", paste0(
journal, ifelse(
is.na(volume),
paste0(", DOI: ", gsub("[.]", " . ", doi)),
paste0(
" ",
volume,
ifelse(is.na(number), "", sprintf("(%s)", number))
)
)
),
type=="inproceedings", booktitle,
type=="incollection", sprintf("Chapter in %s, edited by %s, published by %s", booktitle, editor, publisher),
type=="phdthesis", paste("PHD thesis,", school),
type=="unpublished", note
)][, .(type, year, venue=substr(venue, nchar(venue)-30,nchar(venue)))]
## Key: <type>
## type year venue
## <char> <char> <char>
## 1: article 2021 Functional Ecology 35(4)
## 2: article 2025 BMC Bioinformatics 26(74)
## 3: article 2018 ournal of Human Genetics 103(4)
## 4: article 2022 and Graphical Statistics 31(4)
## 5: article 2024 e Learning and Computation 1(3)
## 6: article 2022 s. Comput.-Hum. Interact. 29(2)
## 7: article 2016 Clinical Cancer Research 22(22)
## 8: article 2018 tional Cancer Institute 110(10)
## 9: article 2018 Scientific data 5(1)
## 10: article 2008 Nature biotechnology 26(6)
## 11: article 2021 ers in Biology and Medicine 130
## 12: article 2010 PLoS one 5(8)
## 13: article 2024 nmental Research Letters 19(11)
## 14: article 2023 ics and Automation Letters 8(8)
## 15: article 2023 achine Learning Research 24(70)
## 16: article 2013 BMC Bioinformatics 14(164)
## 17: article 2013 rnal of Statistical Software 54
## 18: article 2014 Bioinformatics 30(11)
## 19: article 2017 Bioinformatics 33(4)
## 20: article 2019 The R Journal 11(2)
## 21: article 2020 achine Learning Research 21(87)
## 22: article 2021 The R Journal 13(1)
## 23: article 2022 of Statistical Software 101(10)
## 24: article 2023 Computational Statistics 38
## 25: article 2019 Biostatistics 21(4)
## 26: article 2024 and Graphical Statistics 33(4)
## 27: article 2021 BMC Bioinformatics 22(323)
## 28: article 2017 Statistics and Computing 27
## 29: article 2022 logy Methods and Protocols 7(1)
## 30: article 2025 Statistics and Computing 35
## 31: article 2023 of Statistical Software 106(6)
## 32: article 2016 Leukemia 30(7)
## 33: article 2019 and Graphical Statistics 28(2)
## 34: article 2014 Cancer Sci 105(7)
## 35: article 2023 Nature 618
## 36: article 2024 Nature 627(8002)
## 37: article 2022 and Graphical Statistics 31(2)
## 38: incollection 2022 iqi Luo, published by CRC Press
## 39: inproceedings 2022 Transdisciplinary AI (TransAI)
## 40: inproceedings 2022 transdisciplinary AI (TransAI)
## 41: inproceedings 2017 formation Processing Systems 30
## 42: inproceedings 2020 Signals, Systems, and Computers
## 43: inproceedings 2020 Medicine Biology Society (EMBC)
## 44: inproceedings 2011 conference on machine learning
## 45: inproceedings 2013 Proc. 30th ICML
## 46: inproceedings 2015 Proc. 32nd ICML
## 47: inproceedings 2020 cific Symposium on Biocomputing
## 48: inproceedings 2022 Transdisciplinary AI (TransAI)
## 49: inproceedings 2021 Reliability Engineering (ISSRE)
## 50: inproceedings 2023 Technology and Computing (IETC)
## 51: phdthesis 2012 le normale supérieure de Cachan
## 52: unpublished 2025 mputers in Biology and Medicine
## 53: unpublished 2026 In Progress
## 54: unpublished 2025 Under review at R Journal
## 55: unpublished 2024 w at Computational Intelligence
## 56: unpublished 2015 Preprint arXiv:1509.00368
## 57: unpublished 2016 onference, textbook in progress
## 58: unpublished 2017 onference, textbook in progress
## 59: unpublished 2023 In progress
## 60: unpublished 2025 Journal of Statistical Software
## 61: unpublished 2024 Canadian Journal of Statistics
## 62: unpublished 2024 In progress
## 63: unpublished 2024 In progress
## 64: unpublished 2024 stical Analysis and Data Mining
## 65: unpublished 2026 mitted to INSAR 2026 conference
## 66: unpublished 2025 iew at Computational Statistics
## 67: unpublished 2025 iew at Statistics and Computing
## 68: unpublished 2025 enance and Evolution), Sep 2025
## 69: unpublished 2023 al of Machine Learning Research
## 70: unpublished 2025 r review at Academic Pediatrics
## 71: unpublished 2024 Observation and Geoinformation
## 72: unpublished 2024 In progress
## 73: unpublished 2014 Preprint arXiv:1401.8008
## type year venue
## <char> <char> <char>
Authors
Author names come in two forms:
- Family, Given1 Given2
- Given1 Given2 Family
subject <- c("Toby Dylan Hocking", "Hocking, Toby Dylan")
alt.pattern <- nc::alternatives_with_shared_groups(
family="[A-Z][^,]+",
given="[^,]+",
list("^", given, " ", family, "$"),
list("^", family, ", ", given, "$"))
nc::capture_first_vec(subject, alt.pattern)
## given family
## <char> <char>
## 1: Toby Dylan Hocking
## 2: Toby Dylan Hocking
The pattern above matches either of the two forms. Below we use it to match all of the data.
(authors <- refs.wide[, {
complete <- strsplit(author, split=" and ")[[1]]
data.table(complete, nc::capture_first_vec(
complete,
alt.pattern,
nomatch.error=FALSE))
}, by=ref
][
, abbrev := gsub("[a-z. ]", "", given)
][
, show := ifelse(is.na(family), complete, paste(family, abbrev))
][])
## ref complete given family abbrev show
## <char> <char> <char> <char> <char> <char>
## 1: Abraham2021gut Abraham, Andrew J. Andrew J. Abraham AJ Abraham AJ
## 2: Abraham2021gut Prys-Jones, Tomos O. Tomos O. Prys-Jones TO Prys-Jones TO
## 3: Abraham2021gut De Cuyper, Annelies Annelies De Cuyper A De Cuyper A
## 4: Abraham2021gut Ridenour, Chase Chase Ridenour C Ridenour C
## 5: Abraham2021gut Hempson, Gareth P. Gareth P. Hempson GP Hempson GP
## ---
## 417: Truong2024circular Toby Dylan Hocking Toby Dylan Hocking TD Hocking TD
## 418: Venuto2014support Venuto, D D Venuto D Venuto D
## 419: Venuto2014support Hocking, Toby Dylan Toby Dylan Hocking TD Hocking TD
## 420: Venuto2014support Sphanurattana, L L Sphanurattana L Sphanurattana L
## 421: Venuto2014support Sugiyama, M M Sugiyama M Sugiyama M
The table above shows all names standardized to a common format in the show column.
Below we verify that all names matched.
authors[is.na(family)]
## Empty data.table (0 rows and 6 cols): ref,complete,given,family,abbrev,show
The table above shows that there are no entries that did not match the regex, which is OK.
abbrev.dt <- authors[, .(
authors_abbrev=paste(show, collapse=", ")
), by=ref]
abbrev.dt[, length(grep("Hocking",authors_abbrev))]
## [1] 73
The output above shows that there are 73 items for which I am listed as an author.
abbrev.dt[, .(ref, authors_abbrev=substr(authors_abbrev,1,30))]
## ref authors_abbrev
## <char> <char>
## 1: Abraham2021gut Abraham AJ, Prys-Jones TO, De
## 2: Agyapong2025cv Agyapong D, Propster JR, Marks
## 3: Alirezaie2018clinpred Alirezaie N, Kernohan KD, Hart
## 4: Barnwal2022jcgs Barnwal A, Cho H, Hocking T
## 5: Bodine2024mapping Bodine CS, Buscombe D, Hocking
## 6: Chaves2022chatbot Chaves AP, Egbert J, Hocking T
## 7: Chicard2016cancer Chicard M, Boyault S, Colmet D
## 8: Depuydt2018genomic Depuydt P, Boeva V, Hocking TD
## 9: Depuydt2018meta Depuydt P, Koster J, Boeva V,
## 10: Doyon2008heritable Doyon Y, McCammon JM, Miller J
## 11: Fotoohinasab2021greedy Fotoohinasab A, Hocking T, Afg
## 12: Gautier2010bayesian Gautier M, Hocking TD, Foulley
## 13: Gurney2024power Gurney KR, Aslam B, Dass P, Ga
## 14: Harshe2023exoskeleton Harshe K, Williams JR, Hocking
## 15: Hillman2023jmlr Hillman J, Hocking TD
## 16: Hocking2013bioinformatics Hocking TD, Schleiermacher G,
## 17: Hocking2013sustainable Hocking TD, Wutzler T, Ponting
## 18: Hocking2014bioinformatics Hocking TD, Boeva V, Rigaill G
## 19: Hocking2017bioinfo Hocking TD, Goerner-Potvin P,
## 20: Hocking2019regex Hocking TD
## 21: Hocking2020jmlr Hocking TD, Rigaill G, Fearnhe
## 22: Hocking2021reshaping Hocking TD
## 23: Hocking2022jss Hocking TD, Rigaill G, Fearnhe
## 24: Hocking2023lopart Hocking TD, Srivastava A
## 25: Jewell2019biostatistics Jewell SW, Hocking TD, Fearnhe
## 26: Kaufman2024functional Kaufman JM, Stenberg AJ, Hocki
## 27: Liehrmann2021chipseq Liehrmann A, Rigaill G, Hockin
## 28: Maidstone2017optimal Maidstone R, Hocking T, Rigail
## 29: Mihaljevic2022sparsemodr Mihaljevic JR, Borkovec S, Rat
## 30: Nguyen2025mlp Nguyen TL, Hocking TD
## 31: Runge2023jss Runge V, Hocking TD, Romano G,
## 32: Shimada2016leukemia Shimada K, Shimada S, Sugimoto
## 33: Sievert2019jcgs Sievert C, VanderPlas S, Cai J
## 34: Suguro2014cancer Suguro M, Yoshida N, Umino A,
## 35: Tao2023nature Tao F, Huang Y, Hungate BA, Ma
## 36: Tao2024reply Tao F, Houlton BZ, Frey SD, Le
## 37: Vargovich2022breakpoints Vargovich J, Hocking TD
## 38: Hocking2022intro Hocking TD
## 39: Barr2022classifying Barr JR, Hocking TD, Morton G,
## 40: Barr2022graph Barr JR, Shaw P, Abu-Khzam FN,
## 41: Drouin2017mmit Drouin A, Hocking T, Laviolett
## 42: Fotoohinasab2020automaticQRSdetectionAsilomar Fotoohinasab A, Hocking T, Afg
## 43: Fotoohinasab2020segmentationEMBC Fotoohinasab A, Hocking T, Afg
## 44: Hocking2011clusterpath Hocking TD, Joulin A, Bach F,
## 45: Hocking2013icml Rigaill G, Hocking T, Vert J-P
## 46: Hocking2015icml Hocking TD, Rigaill G, Bourque
## 47: Hocking2020psb Hocking TD, Bourque G
## 48: Hocking2022interpretable Hocking TD, Barr JR, Thatcher
## 49: Kolla2021fuzz Kolla AC, Groce A, Hocking TD
## 50: Sweeney2023insect Sweeney N, Xu C, Shaw JA, Hock
## 51: Hocking2012phd Hocking TD
## 52: Agyapong2026fused Agyapong D, Beatty BH, Kennedy
## 53: Agyapong2026poisson Agyapong D, Chiquet J, Marks J
## 54: Amoakohene2025asymptotic Amoakohene D, Chetia A, Steinm
## 55: Fowler2024line Fowler J, Hocking TD
## 56: Hocking2015breakpointError Hocking TD
## 57: Hocking2016interactive Hocking TD, Ekstrøm CT
## 58: Hocking2017changepoint Hocking TD, Killick R
## 59: Hocking2023functional Hocking TD
## 60: Hocking2024binsegRcpp Hocking TD
## 61: Hocking2024finite Hocking TD
## 62: Hocking2024hmm Hocking TD
## 63: Hocking2024mlr3resampling Hocking TD
## 64: Hocking2024soak Bodine CS, Thibault G, Arellan
## 65: Lindly2026autism Lindly O, Zuckerman K, Hocking
## 66: Nguyen2025automatic Nguyen TL, Hocking TD
## 67: Nguyen2025comparative Nguyen TL, Hocking TD
## 68: Oliveira2025governance Oliveira P, Amoakohene D, Hock
## 69: Rust2023pairs Rust KR, Hocking TD
## 70: Sutherland2025autism Sutherland V, Hocking TD, Folc
## 71: Thibault2024forest Thibault G, Morin-Bernard A, S
## 72: Truong2024circular Truong C, Hocking TD
## 73: Venuto2014support Venuto D, Hocking TD, Sphanura
## ref authors_abbrev
## <char> <char>
The output above shows the abbreviated author list is reasonable.
Count article types
type2long <- c(
article="journal paper",
incollection="book chapter",
inproceedings="conference paper",
phdthesis="PHD thesis",
unpublished="in progress")
refs.wide[, .(count=.N), by=.(Type=type2long[type])][order(-count)]
## Type count
## <char> <int>
## 1: journal paper 37
## 2: in progress 22
## 3: conference paper 12
## 4: book chapter 1
## 5: PHD thesis 1
Output markdown
The code below joins the authors back to the original table, then outputs markdown.
abbrev.wide <- refs.wide[
abbrev.dt, on="ref"
][, let(
heading = ifelse(type=="unpublished", "In progress", year),
citation = sprintf("- %s (%s). %s. %s. %s", authors_abbrev, year, title, venue, links)
)][order(-heading, -year, authors_abbrev)]
abbrev.some <- abbrev.wide[unique(heading)[1:3], .SD[1:2], on="heading", by=heading]
abbrev.some <- abbrev.wide
abbrev.some[
, .(markdown=sprintf("### %s\n%s\n", heading, paste(citation, collapse="\n")))
, by=heading
][
, cat(paste(markdown, collapse="\n"))
]
In progress
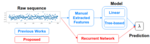
- Agyapong D, Chiquet J, Marks J, Hocking TD (2026). Identifying regimes where graphical models out-perform linear models for microbiome counts. In Progress. Reproducible
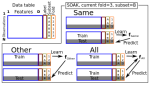
- Lindly O, Zuckerman K, Hocking TD, Folch DC, Sutherland V, Curry T (2026). Predict, Profile, Prioritize: Machine Learning Reveals Hidden Patterns in Autism Service Access Disparities. Abstract submitted to INSAR 2026 conference. Reproducible
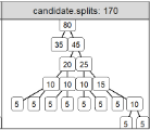
- Agyapong D, Beatty BH, Kennedy PG, Marks JC, Hocking TD (2025). Fused Lasso Improves Accuracy of Co-occurrence Network Inference in Grouped Samples. Preprint arXiv:2509.09413, under review at Computers in Biology and Medicine. Reproducible, Software
- Amoakohene D, Chetia A, Steinmacher I, Hocking TD (2025). Asymptotic benchmarking using the atime package. Under review at R Journal. Reproducible, Software
- Hocking TD (2025). Comparing binsegRcpp with other implementations of binary segmentation for changepoint detection. Under review at Journal of Statistical Software. Preprint, Reproducible, Software
- Nguyen TL, Hocking TD (2025). Penalty Learning for Optimal Partitioning via Automatic Feature Extraction. Preprint arXiv:2505.07413, under review at Computational Statistics. Preprint, Reproducible
- Nguyen TL, Hocking TD (2025). Interval Regression: A Comparative Study with Proposed Models. Preprint arXiv:2503.02011, under review at Statistics and Computing. Preprint, Reproducible
- Oliveira P, Amoakohene D, Hocking TD, Gerosa M, Steinmacher I (2025). Governance Matters: Lessons from Restructuring the data.table OSS Project. ICSME (International Conference on Software Maintenance and Evolution), Sep 2025. Abstract
- Sutherland V, Hocking TD, Folch DC, Underwood Carrasco VI, Zuckerman KE, Lindly OJ (2025). Autism Diagnostic Determinations of Primary Care Providers from 2013 to 2023. Under review at Academic Pediatrics. Related poster Trends in Autism Diagnostic Determinations by Primary Care Providers Among U.S. Children from 2013 to 2023: Effects of Child Age, Race and Ethnicity, and Poverty Level was selected as one of the Top-Rated Abstracts at INSAR 2025 conference.
- Bodine CS, Thibault G, Arellano PN, Shenkin AF, Lindly O, Hocking TD (2024). SOAK: Same/Other/All K-fold cross-validation for estimating similarity of patterns in data subsets. Preprint arXiv:2410.08643, under review at Statistical Analysis and Data Mining. Preprint, Software, Reproducible
- Fowler J, Hocking TD (2024). Efficient line search for optimizing Area Under the ROC Curve in gradient descent. Preprint arXiv:2410.08635, under review at Computational Intelligence. Preprint, Talk announcement for JSM’23 Toronto, Video of talk at Université Laval July 2023, Slides PDF, source
- Hocking TD (2024). Finite Sample Complexity Analysis of Binary Segmentation. Preprint arXiv:2410.08654, under review at Canadian Journal of Statistics. Preprint, Reproducible, Software
- Hocking TD (2024). Teaching Hidden Markov Models Using Interactive Data Visualization. In progress. Reproducible, Slides
- Hocking TD (2024). mlr3resampling: an R implementation of cross-validation for comparing models learned using different train subsets. In progress. Software
- Thibault G, Morin-Bernard A, Sylvain J-D, Drolet G, Roussel J-R, Hocking TD, Achim A (2024). Spatial characterization of burn severity in a boreal forest using high-resolution satellite imagery. Under review at International Journal of Applied Earth Observation and Geoinformation. NA
- Truong C, Hocking TD (2024). Efficient change-point detection for multivariate circular data. In progress. Reproducible, Software
- Hocking TD (2023). Why does functional pruning yield such fast algorithms for optimal changepoint detection?. In progress. Invited talk for TRIPODS seminar video, slides, IEEE NJACS, ASU West ML Day
- Rust KR, Hocking TD (2023). A Log-linear Gradient Descent Algorithm for Unbalanced Binary Classification using the All Pairs Squared Hinge Loss. Preprint arXiv:2302.11062, under review at Journal of Machine Learning Research. Preprint
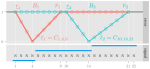
- Hocking TD, Killick R (2017). Introduction to optimal changepoint detection algorithms. Tutorial at international useR 2017 conference, textbook in progress. Conference, Book, Reproducible
- Hocking TD, Ekstrøm CT (2016). Understanding and Creating Interactive Graphics. Tutorial at international useR 2016 conference, textbook in progress. Conference, Manual, Reproducible
- Hocking TD (2015). A breakpoint detection error function for segmentation model selection and validation. Preprint arXiv:1509.00368. Preprint, Software, Reproducible
- Venuto D, Hocking TD, Sphanurattana L, Sugiyama M (2014). Support vector comparison machines. Preprint arXiv:1401.8008. Preprint, Software, Reproducible
2025
- Agyapong D, Propster JR, Marks J, Hocking TD (2025). Cross-Validation for Training and Testing Co-occurrence Network Inference Algorithms. BMC Bioinformatics 26(74). Publisher, Preprint, Reproducible
- Nguyen TL, Hocking TD (2025). Penalty Learning for Optimal Partitioning using Multilayer Perceptron. Statistics and Computing 35. Publisher, Preprint, Reproducible
2024
- Bodine CS, Buscombe D, Hocking TD (2024). Automated River Substrate Mapping From Sonar Imagery With Machine Learning. Journal of Geophysical Research: Machine Learning and Computation 1(3). DOI, Preprint, Software
- Gurney KR, Aslam B, Dass P, Gawuc L, Hocking TD, Barber JJ, Kato A (2024). Assessment of the Climate Trace global powerplant CO2 emissions. Environmental Research Letters 19(11). Publisher
- Kaufman JM, Stenberg AJ, Hocking TD (2024). Functional Labeled Optimal Partitioning. Journal of Computational and Graphical Statistics 33(4). DOI, Preprint, Software, Reproducible, Preliminary code
- Tao F, Houlton BZ, Frey SD, Lehmann J, Manzoni S, Huang Y, Jiang L, Mishra U, Hungate BA, Schmidt MWI, Reichstein M, Carvalhais N, Ciais P, Wang Y-P, Ahrens B, Hugelius G, Hocking TD, Lu X, Shi Z, Viatkin K, Vargas R, Yigini Y, Omuto C, Malik AA, Peralta G, Cuevas-Corona R, Paolo LED, Luotto I, Liao C, Liang Y-S, Saynes VS, Huang X, Luo Y (2024). Reply to: Model uncertainty obscures major driver of soil carbon. Nature 627(8002). Publisher, Preprint
2023
- Harshe K, Williams JR, Hocking TD, Lerner ZF (2023). Predicting Neuromuscular Engagement to Improve Gait Training With a Robotic Ankle Exoskeleton. IEEE Robotics and Automation Letters 8(8). Publisher
- Hillman J, Hocking TD (2023). Optimizing ROC Curves with a Sort-Based Surrogate Loss for Binary Classification and Changepoint Detection. Journal of Machine Learning Research 24(70). Publisher, Preprint, Software, Reproducible, Video
- Hocking TD, Srivastava A (2023). Labeled optimal partitioning. Computational Statistics 38. Publisher, Preprint, Video, Software, Reproducible
- Runge V, Hocking TD, Romano G, Afghah F, Fearnhead P, Rigaill G (2023). gfpop: An R Package for Univariate Graph-Constrained Change-Point Detection. Journal of Statistical Software 106(6). Publisher, Preprint, Software, GUI, Reproducible
- Sweeney N, Xu C, Shaw JA, Hocking TD, Whitaker BM (2023). Insect Identification in Pulsed Lidar Images Using Changepoint Detection Algorithms. 2023 Intermountain Engineering, Technology and Computing (IETC). Publisher
- Tao F, Huang Y, Hungate BA, Manzoni S, Frey SD, Schmidt MWI, Reichstein M, Carvalhais N, Ciais P, Jiang L, Lehmann J, Mishra U, Hugelius G, Hocking TD, Lu X, Shi Z, Viatkin K, Vargas R, Yigini Y, Omuto C, Malik AA, Perualta G, Cuevas-Corona R, Paolo LED, Luotto I, Liao C, Liang Y-S, Saynes VS, Huang X, Luo Y (2023). Microbial carbon use efficiency promotes global soil carbon storage. Nature 618. Publisher
2022
- Barnwal A, Cho H, Hocking T (2022). Survival Regression with Accelerated Failure Time Model in XGBoost. Journal of Computational and Graphical Statistics 31(4). Publisher, Preprint, Software, Documentation, Video, Reproducible
- Barr JR, Hocking TD, Morton G, Thatcher T, Shaw P (2022). Classifying Imbalanced Data with AUM Loss. 2022 Fourth International Conference on Transdisciplinary AI (TransAI). Publisher
- Barr JR, Shaw P, Abu-Khzam FN, Thatcher T, Hocking TD (2022). Graph embedding: A methodological survey. 2022 fourth international conference on transdisciplinary AI (TransAI). Publisher
- Chaves AP, Egbert J, Hocking T, Doerry E, Gerosa MA (2022). Chatbots Language Design: The Influence of Language Variation on User Experience with Tourist Assistant Chatbots. ACM Trans. Comput.-Hum. Interact. 29(2). Publisher, Preprint
- Hocking TD (2022). Introduction to machine learning and neural networks. Chapter in Land Carbon Cycle Modeling: Matrix Approach, Data Assimilation, and Ecological Forecasting, edited by Yiqi Luo, published by CRC Press. Publisher, My chapter, Reproducible, Video
- Hocking TD, Barr JR, Thatcher T (2022). Interpretable linear models for predicting security vulnerabilities in source code. 2022 Fourth International Conference on Transdisciplinary AI (TransAI). Publisher
- Hocking TD, Rigaill G, Fearnhead P, Bourque G (2022). Generalized Functional Pruning Optimal Partitioning (GFPOP) for Constrained Changepoint Detection in Genomic Data. Journal of Statistical Software 101(10). Publisher, Software, Reproducible, Slides, Video
- Mihaljevic JR, Borkovec S, Ratnavale S, Hocking TD, Banister KE, Eppinger JE, Hepp C, Doerry E (2022). SPARSEMODr: Rapidly simulate spatially explicit and stochastic models of COVID-19 and other infectious diseases. Biology Methods and Protocols 7(1). Publisher, Preprint, Software
- Vargovich J, Hocking TD (2022). Linear time dynamic programming for computing breakpoints in the regularization path of models selected from a finite set. Journal of Computational and Graphical Statistics 31(2). Publisher, Preprint, Software, Reproducible
2021
- Abraham AJ, Prys-Jones TO, De Cuyper A, Ridenour C, Hempson GP, Hocking T, Clauss M, Doughty CE (2021). Improved estimation of gut passage time considerably affects trait-based dispersal models. Functional Ecology 35(4). Publisher
- Fotoohinasab A, Hocking T, Afghah F (2021). A greedy graph search algorithm based on changepoint analysis for automatic QRS complex detection. Computers in Biology and Medicine 130. Publisher, Preprint
- Hocking TD (2021). Wide-to-tall Data Reshaping Using Regular Expressions and the nc Package. The R Journal 13(1). Publisher, Software, Reproducible
- Kolla AC, Groce A, Hocking TD (2021). Fuzz Testing the Compiled Code in R Packages. 2021 IEEE 32nd International Symposium on Software Reliability Engineering (ISSRE). Publisher, Software, GitHub Action, Abstract, Video, Blog, Results
- Liehrmann A, Rigaill G, Hocking TD (2021). Increased peak detection accuracy in over-dispersed ChIP-seq data with supervised segmentation models. BMC Bioinformatics 22(323). Publisher, Reproducible, Software
2020
- Fotoohinasab A, Hocking T, Afghah F (2020). A Graph-Constrained Changepoint Learning Approach for Automatic QRS-Complex Detection. 2020 54th Asilomar Conference on Signals, Systems, and Computers. Publisher, Abstract, Preprint
- Fotoohinasab A, Hocking T, Afghah F (2020). A Graph-constrained Changepoint Detection Approach for ECG Segmentation. 2020 42nd Annual International Conference of the IEEE Engineering in Medicine Biology Society (EMBC). Publisher
- Hocking TD, Bourque G (2020). Machine Learning Algorithms for Simultaneous Supervised Detection of Peaks in Multiple Samples and Cell Types. Proc. Pacific Symposium on Biocomputing. Publisher, Software, Preprint, Reproducible
- Hocking TD, Rigaill G, Fearnhead P, Bourque G (2020). Constrained Dynamic Programming and Supervised Penalty Learning Algorithms for Peak Detection in Genomic Data. Journal of Machine Learning Research 21(87). Publisher, Preprint, Software, Reproducible
2019
- Hocking TD (2019). Comparing namedCapture with other R packages for regular expressions. The R Journal 11(2). Publisher, Software, Reproducible
- Jewell SW, Hocking TD, Fearnhead P, Witten DM (2019). Fast nonconvex deconvolution of calcium imaging data. Biostatistics 21(4). Pubmed
- Sievert C, VanderPlas S, Cai J, Ferris K, Khan FUF, Hocking TD (2019). Extending ggplot2 for Linked and Animated Web Graphics. Journal of Computational and Graphical Statistics 28(2). Publisher, Software, Reproducible, Interactive Figures
2018
- Alirezaie N, Kernohan KD, Hartley T, Majewski J, Hocking TD (2018). ClinPred: Prediction Tool to Identify Disease-Relevant Nonsynonymous Single-Nucleotide Variants. The American Journal of Human Genetics 103(4). DOI, Software, used in dbNSFP
- Depuydt P, Boeva V, Hocking TD, Cannoodt R, Ambros IM, Ambros PF, Asgharzadeh S, Attiyeh EF, Combaret Vé, Defferrari R, Fischer M, Hero B, Hogarty MD, Irwin MS, Koster J, Kreissman S, Ladenstein R, Lapouble E, Laureys Gè, London WB, Mazzocco K, Nakagawara A, Noguera R, Ohira M, Park JR, Pötschger U, Theissen J, Tonini GP, Valteau-Couanet D, Varesio L, Versteeg R, Speleman F, Maris JM, Schleiermacher G, Preter KD (2018). Genomic amplifications and distal 6q loss: novel markers for poor survival in high-risk neuroblastoma patients. JNCI: Journal of the National Cancer Institute 110(10). Publisher
- Depuydt P, Koster J, Boeva V, Hocking TD, Speleman F, Schleiermacher G, De Preter K (2018). Meta-mining of copy number profiles of high-risk neuroblastoma tumors. Scientific data 5(1). Publisher
2017
- Drouin A, Hocking T, Laviolette F (2017). Maximum Margin Interval Trees. Advances in Neural Information Processing Systems 30. Publisher, Software, Reproducible, Preprint, Video
- Hocking TD, Goerner-Potvin P, Morin A, Shao X, Pastinen T, Bourque G (2017). Optimizing ChIP-seq peak detectors using visual labels and supervised machine learning. Bioinformatics 33(4). Pubmed, Software, Reproducible, Data
- Maidstone R, Hocking T, Rigaill G, Fearnhead P (2017). On optimal multiple changepoint algorithms for large data. Statistics and Computing 27. Publisher, Software, Reproducible
2016
- Chicard M, Boyault S, Colmet Daage L, Richer W, Gentien D, Pierron G, Lapouble E, Bellini A, Clement N, Iacono I, Bréjon Sé, Carrere M, Reyes Cé, Hocking T, Bernard V, Peuchmaur M, Corradini Nè, Faure-Conter Cé, Coze C, Plantaz D, Defachelles AS, Thebaud E, Gambart M, Millot Féé, Valteau-Couanet D, Michon J, Puisieux A, Delattre O, Combaret Vé, Schleiermacher G (2016). Genomic Copy Number Profiling Using Circulating Free Tumor DNA Highlights Heterogeneity in Neuroblastoma. Clinical Cancer Research 22(22). Publisher
- Shimada K, Shimada S, Sugimoto K, Nakatochi M, Suguro M, Hirakawa A, Hocking TD, Takeuchi I, Tokunaga T, Takagi Y, Sakamoto A, Aoki T, Naoe T, Nakamura S, Hayakawa F, Seto M, Tomita A, Kiyoi H (2016). Development and analysis of patient-derived xenograft mouse models in intravascular large B-cell lymphoma. Leukemia 30(7). Pubmed
2015
- Hocking TD, Rigaill G, Bourque G (2015). PeakSeg: constrained optimal segmentation and supervised penalty learning for peak detection in count data. Proc. 32nd ICML. Publisher, Video, Software, Reproducible
2014
- Hocking TD, Boeva V, Rigaill G, Schleiermacher G, Janoueix-Lerosey I, Delattre O, Richer W, Bourdeaut F, Suguro M, Seto M, Bach F, Vert JP (2014). SegAnnDB: interactive Web-based genomic segmentation. Bioinformatics 30(11). Pubmed, Software, Reproducible, Preprint, Package, Reproducible
- Suguro M, Yoshida N, Umino A, Kato H, Tagawa H, Nakagawa M, Fukuhara N, Karnan S, Takeuchi I, Hocking TD, Arita K, Karube K, Tsuzuki S, Nakamura S, Kinoshita T, Seto M (2014). Clonal heterogeneity of lymphoid malignancies correlates with poor prognosis. Cancer Sci 105(7). Pubmed
2013
- Hocking TD, Schleiermacher G, Janoueix-Lerosey I, Boeva V, Cappo J, Delattre O, Bach F, Vert J-P (2013). Learning smoothing models of copy number profiles using breakpoint annotations. BMC Bioinformatics 14(164). Publisher, Software, Reproducible
- Hocking TD, Wutzler T, Ponting K, Grosjean P (2013). Sustainable, Extensible Documentation Generation using inlinedocs. Journal of Statistical Software 54. Publisher, Software, Reproducible
- Rigaill G, Hocking T, Vert J-P, Bach F (2013). Learning sparse penalties for change-point detection using max margin interval regression. Proc. 30th ICML. Publisher, Video, Software, Reproducible
2012
- Hocking TD (2012). Learning algorithms and statistical software, with applications to bioinformatics. PHD thesis, Ecole normale supérieure de Cachan. Publisher, Reproducible
2011
- Hocking TD, Joulin A, Bach F, Vert J-P (2011). Clusterpath an algorithm for clustering using convex fusion penalties. 28th international conference on machine learning. Publisher, Video, Software, Reproducible, Cited in Ihaka lecture
2010
- Gautier M, Hocking TD, Foulley J-L (2010). A Bayesian outlier criterion to detect SNPs under selection in large data sets. PLoS one 5(8). Publisher
2008
- Doyon Y, McCammon JM, Miller JC, Faraji F, Ngo C, Katibah GE, Amora R, Hocking TD, Zhang L, Rebar EJ, Gregory PD, Urnov FD, Amacher SL (2008). Heritable targeted gene disruption in zebrafish using designed zinc-finger nucleases. Nature biotechnology 26(6). Pubmed NULL
Another output: markdown table with images
One advantage of this is that we can easily modify output formats.
for(large.png in Sys.glob("../assets/img/publications/*.png")){
thumb.png <- file.path(
dirname(large.png),
"thumb",
basename(large.png))
if(!file.exists(thumb.png)){
dir.create(dirname(thumb.png))
convert.cmd <- paste("convert", large.png, "-scale 150", thumb.png)
print(convert.cmd)
system(convert.cmd)
}
}
some.out <- abbrev.some[
, figure.png := sprintf("/assets/img/publications/thumb/%s.png", ref)
][, .(
Figure=ifelse(
file.exists(file.path("..", figure.png)),
sprintf('<img src="%s" width="150" />', figure.png),
""),
Published=heading,
Authors=authors_abbrev,
Title=title,
Venue=venue,
Links=ifelse(is.na(links), "", links)
)]
knitr::kable(some.out)
| Figure | Published | Authors | Title | Venue | Links |
|---|---|---|---|---|---|
 |
In progress | Agyapong D, Chiquet J, Marks J, Hocking TD | Identifying regimes where graphical models out-perform linear models for microbiome counts | In Progress | Reproducible |
 |
In progress | Lindly O, Zuckerman K, Hocking TD, Folch DC, Sutherland V, Curry T | Predict, Profile, Prioritize: Machine Learning Reveals Hidden Patterns in Autism Service Access Disparities | Abstract submitted to INSAR 2026 conference | Reproducible |
 |
In progress | Agyapong D, Beatty BH, Kennedy PG, Marks JC, Hocking TD | Fused Lasso Improves Accuracy of Co-occurrence Network Inference in Grouped Samples | Preprint arXiv:2509.09413, under review at Computers in Biology and Medicine | Reproducible, Software |
 |
In progress | Amoakohene D, Chetia A, Steinmacher I, Hocking TD | Asymptotic benchmarking using the atime package | Under review at R Journal | Reproducible, Software |
 |
In progress | Hocking TD | Comparing binsegRcpp with other implementations of binary segmentation for changepoint detection | Under review at Journal of Statistical Software | Preprint, Reproducible, Software |
 |
In progress | Nguyen TL, Hocking TD | Penalty Learning for Optimal Partitioning via Automatic Feature Extraction | Preprint arXiv:2505.07413, under review at Computational Statistics | Preprint, Reproducible |
 |
In progress | Nguyen TL, Hocking TD | Interval Regression: A Comparative Study with Proposed Models | Preprint arXiv:2503.02011, under review at Statistics and Computing | Preprint, Reproducible |
 |
In progress | Oliveira P, Amoakohene D, Hocking TD, Gerosa M, Steinmacher I | Governance Matters: Lessons from Restructuring the data.table OSS Project | ICSME (International Conference on Software Maintenance and Evolution), Sep 2025 | Abstract |
 |
In progress | Sutherland V, Hocking TD, Folch DC, Underwood Carrasco VI, Zuckerman KE, Lindly OJ | Autism Diagnostic Determinations of Primary Care Providers from 2013 to 2023 | Under review at Academic Pediatrics | Related poster Trends in Autism Diagnostic Determinations by Primary Care Providers Among U.S. Children from 2013 to 2023: Effects of Child Age, Race and Ethnicity, and Poverty Level was selected as one of the Top-Rated Abstracts at INSAR 2025 conference. |
 |
In progress | Bodine CS, Thibault G, Arellano PN, Shenkin AF, Lindly O, Hocking TD | SOAK: Same/Other/All K-fold cross-validation for estimating similarity of patterns in data subsets | Preprint arXiv:2410.08643, under review at Statistical Analysis and Data Mining | Preprint, Software, Reproducible |
 |
In progress | Fowler J, Hocking TD | Efficient line search for optimizing Area Under the ROC Curve in gradient descent | Preprint arXiv:2410.08635, under review at Computational Intelligence | Preprint, Talk announcement for JSM’23 Toronto, Video of talk at Université Laval July 2023, Slides PDF, source |
 |
In progress | Hocking TD | Finite Sample Complexity Analysis of Binary Segmentation | Preprint arXiv:2410.08654, under review at Canadian Journal of Statistics | Preprint, Reproducible, Software |
 |
In progress | Hocking TD | Teaching Hidden Markov Models Using Interactive Data Visualization | In progress | Reproducible, Slides |
 |
In progress | Hocking TD | mlr3resampling: an R implementation of cross-validation for comparing models learned using different train subsets | In progress | Software |
 |
In progress | Thibault G, Morin-Bernard A, Sylvain J-D, Drolet G, Roussel J-R, Hocking TD, Achim A | Spatial characterization of burn severity in a boreal forest using high-resolution satellite imagery | Under review at International Journal of Applied Earth Observation and Geoinformation | |
 |
In progress | Truong C, Hocking TD | Efficient change-point detection for multivariate circular data | In progress | Reproducible, Software |
 |
In progress | Hocking TD | Why does functional pruning yield such fast algorithms for optimal changepoint detection? | In progress | Invited talk for TRIPODS seminar video, slides, IEEE NJACS, ASU West ML Day |
 |
In progress | Rust KR, Hocking TD | A Log-linear Gradient Descent Algorithm for Unbalanced Binary Classification using the All Pairs Squared Hinge Loss | Preprint arXiv:2302.11062, under review at Journal of Machine Learning Research | Preprint |
 |
In progress | Hocking TD, Killick R | Introduction to optimal changepoint detection algorithms | Tutorial at international useR 2017 conference, textbook in progress | Conference, Book, Reproducible |
 |
In progress | Hocking TD, Ekstrøm CT | Understanding and Creating Interactive Graphics | Tutorial at international useR 2016 conference, textbook in progress | Conference, Manual, Reproducible |
 |
In progress | Hocking TD | A breakpoint detection error function for segmentation model selection and validation | Preprint arXiv:1509.00368 | Preprint, Software, Reproducible |
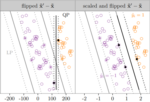 |
In progress | Venuto D, Hocking TD, Sphanurattana L, Sugiyama M | Support vector comparison machines | Preprint arXiv:1401.8008 | Preprint, Software, Reproducible |
 |
2025 | Agyapong D, Propster JR, Marks J, Hocking TD | Cross-Validation for Training and Testing Co-occurrence Network Inference Algorithms | BMC Bioinformatics 26(74) | Publisher, Preprint, Reproducible |
 |
2025 | Nguyen TL, Hocking TD | Penalty Learning for Optimal Partitioning using Multilayer Perceptron | Statistics and Computing 35 | Publisher, Preprint, Reproducible |
 |
2024 | Bodine CS, Buscombe D, Hocking TD | Automated River Substrate Mapping From Sonar Imagery With Machine Learning | Journal of Geophysical Research: Machine Learning and Computation 1(3) | DOI, Preprint, Software |
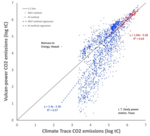 |
2024 | Gurney KR, Aslam B, Dass P, Gawuc L, Hocking TD, Barber JJ, Kato A | Assessment of the Climate Trace global powerplant CO2 emissions | Environmental Research Letters 19(11) | Publisher |
 |
2024 | Kaufman JM, Stenberg AJ, Hocking TD | Functional Labeled Optimal Partitioning | Journal of Computational and Graphical Statistics 33(4) | DOI, Preprint, Software, Reproducible, Preliminary code |
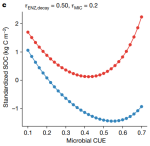 |
2024 | Tao F, Houlton BZ, Frey SD, Lehmann J, Manzoni S, Huang Y, Jiang L, Mishra U, Hungate BA, Schmidt MWI, Reichstein M, Carvalhais N, Ciais P, Wang Y-P, Ahrens B, Hugelius G, Hocking TD, Lu X, Shi Z, Viatkin K, Vargas R, Yigini Y, Omuto C, Malik AA, Peralta G, Cuevas-Corona R, Paolo LED, Luotto I, Liao C, Liang Y-S, Saynes VS, Huang X, Luo Y | Reply to: Model uncertainty obscures major driver of soil carbon | Nature 627(8002) | Publisher, Preprint |
 |
2023 | Harshe K, Williams JR, Hocking TD, Lerner ZF | Predicting Neuromuscular Engagement to Improve Gait Training With a Robotic Ankle Exoskeleton | IEEE Robotics and Automation Letters 8(8) | Publisher |
 |
2023 | Hillman J, Hocking TD | Optimizing ROC Curves with a Sort-Based Surrogate Loss for Binary Classification and Changepoint Detection | Journal of Machine Learning Research 24(70) | Publisher, Preprint, Software, Reproducible, Video |
 |
2023 | Hocking TD, Srivastava A | Labeled optimal partitioning | Computational Statistics 38 | Publisher, Preprint, Video, Software, Reproducible |
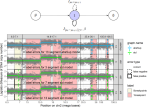 |
2023 | Runge V, Hocking TD, Romano G, Afghah F, Fearnhead P, Rigaill G | gfpop: An R Package for Univariate Graph-Constrained Change-Point Detection | Journal of Statistical Software 106(6) | Publisher, Preprint, Software, GUI, Reproducible |
 |
2023 | Sweeney N, Xu C, Shaw JA, Hocking TD, Whitaker BM | Insect Identification in Pulsed Lidar Images Using Changepoint Detection Algorithms | 2023 Intermountain Engineering, Technology and Computing (IETC) | Publisher |
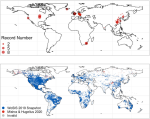 |
2023 | Tao F, Huang Y, Hungate BA, Manzoni S, Frey SD, Schmidt MWI, Reichstein M, Carvalhais N, Ciais P, Jiang L, Lehmann J, Mishra U, Hugelius G, Hocking TD, Lu X, Shi Z, Viatkin K, Vargas R, Yigini Y, Omuto C, Malik AA, Perualta G, Cuevas-Corona R, Paolo LED, Luotto I, Liao C, Liang Y-S, Saynes VS, Huang X, Luo Y | Microbial carbon use efficiency promotes global soil carbon storage | Nature 618 | Publisher |
 |
2022 | Barnwal A, Cho H, Hocking T | Survival Regression with Accelerated Failure Time Model in XGBoost | Journal of Computational and Graphical Statistics 31(4) | Publisher, Preprint, Software, Documentation, Video, Reproducible |
 |
2022 | Barr JR, Hocking TD, Morton G, Thatcher T, Shaw P | Classifying Imbalanced Data with AUM Loss | 2022 Fourth International Conference on Transdisciplinary AI (TransAI) | Publisher |
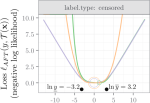
 |
2022 | Barr JR, Shaw P, Abu-Khzam FN, Thatcher T, Hocking TD | Graph embedding: A methodological survey | 2022 fourth international conference on transdisciplinary AI (TransAI) | Publisher |
 |
2022 | Chaves AP, Egbert J, Hocking T, Doerry E, Gerosa MA | Chatbots Language Design: The Influence of Language Variation on User Experience with Tourist Assistant Chatbots | ACM Trans. Comput.-Hum. Interact. 29(2) | Publisher, Preprint |
 |
2022 | Hocking TD | Introduction to machine learning and neural networks | Chapter in Land Carbon Cycle Modeling: Matrix Approach, Data Assimilation, and Ecological Forecasting, edited by Yiqi Luo, published by CRC Press | Publisher, My chapter, Reproducible, Video |
 |
2022 | Hocking TD, Barr JR, Thatcher T | Interpretable linear models for predicting security vulnerabilities in source code | 2022 Fourth International Conference on Transdisciplinary AI (TransAI) | Publisher |
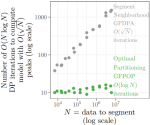 |
2022 | Hocking TD, Rigaill G, Fearnhead P, Bourque G | Generalized Functional Pruning Optimal Partitioning (GFPOP) for Constrained Changepoint Detection in Genomic Data | Journal of Statistical Software 101(10) | Publisher, Software, Reproducible, Slides, Video |
 |
2022 | Mihaljevic JR, Borkovec S, Ratnavale S, Hocking TD, Banister KE, Eppinger JE, Hepp C, Doerry E | SPARSEMODr: Rapidly simulate spatially explicit and stochastic models of COVID-19 and other infectious diseases | Biology Methods and Protocols 7(1) | Publisher, Preprint, Software |
 |
2022 | Vargovich J, Hocking TD | Linear time dynamic programming for computing breakpoints in the regularization path of models selected from a finite set | Journal of Computational and Graphical Statistics 31(2) | Publisher, Preprint, Software, Reproducible |
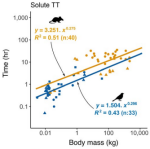 |
2021 | Abraham AJ, Prys-Jones TO, De Cuyper A, Ridenour C, Hempson GP, Hocking T, Clauss M, Doughty CE | Improved estimation of gut passage time considerably affects trait-based dispersal models | Functional Ecology 35(4) | Publisher |
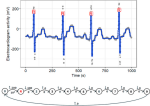 |
2021 | Fotoohinasab A, Hocking T, Afghah F | A greedy graph search algorithm based on changepoint analysis for automatic QRS complex detection | Computers in Biology and Medicine 130 | Publisher, Preprint |
 |
2021 | Hocking TD | Wide-to-tall Data Reshaping Using Regular Expressions and the nc Package | The R Journal 13(1) | Publisher, Software, Reproducible |
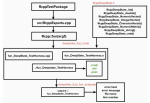 |
2021 | Kolla AC, Groce A, Hocking TD | Fuzz Testing the Compiled Code in R Packages | 2021 IEEE 32nd International Symposium on Software Reliability Engineering (ISSRE) | Publisher, Software, GitHub Action, Abstract, Video, Blog, Results |
 |
2021 | Liehrmann A, Rigaill G, Hocking TD | Increased peak detection accuracy in over-dispersed ChIP-seq data with supervised segmentation models | BMC Bioinformatics 22(323) | Publisher, Reproducible, Software |
 |
2020 | Fotoohinasab A, Hocking T, Afghah F | A Graph-Constrained Changepoint Learning Approach for Automatic QRS-Complex Detection | 2020 54th Asilomar Conference on Signals, Systems, and Computers | Publisher, Abstract, Preprint |
 |
2020 | Fotoohinasab A, Hocking T, Afghah F | A Graph-constrained Changepoint Detection Approach for ECG Segmentation | 2020 42nd Annual International Conference of the IEEE Engineering in Medicine Biology Society (EMBC) | Publisher |
 |
2020 | Hocking TD, Bourque G | Machine Learning Algorithms for Simultaneous Supervised Detection of Peaks in Multiple Samples and Cell Types | Proc. Pacific Symposium on Biocomputing | Publisher, Software, Preprint, Reproducible |
 |
2020 | Hocking TD, Rigaill G, Fearnhead P, Bourque G | Constrained Dynamic Programming and Supervised Penalty Learning Algorithms for Peak Detection in Genomic Data | Journal of Machine Learning Research 21(87) | Publisher, Preprint, Software, Reproducible |
 |
2019 | Hocking TD | Comparing namedCapture with other R packages for regular expressions | The R Journal 11(2) | Publisher, Software, Reproducible |
 |
2019 | Jewell SW, Hocking TD, Fearnhead P, Witten DM | Fast nonconvex deconvolution of calcium imaging data | Biostatistics 21(4) | Pubmed |
 |
2019 | Sievert C, VanderPlas S, Cai J, Ferris K, Khan FUF, Hocking TD | Extending ggplot2 for Linked and Animated Web Graphics | Journal of Computational and Graphical Statistics 28(2) | Publisher, Software, Reproducible, Interactive Figures |
 |
2018 | Alirezaie N, Kernohan KD, Hartley T, Majewski J, Hocking TD | ClinPred: Prediction Tool to Identify Disease-Relevant Nonsynonymous Single-Nucleotide Variants | The American Journal of Human Genetics 103(4) | DOI, Software, used in dbNSFP |
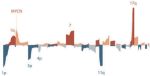 |
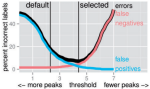
2018 | Depuydt P, Boeva V, Hocking TD, Cannoodt R, Ambros IM, Ambros PF, Asgharzadeh S, Attiyeh EF, Combaret Vé, Defferrari R, Fischer M, Hero B, Hogarty MD, Irwin MS, Koster J, Kreissman S, Ladenstein R, Lapouble E, Laureys Gè, London WB, Mazzocco K, Nakagawara A, Noguera R, Ohira M, Park JR, Pötschger U, Theissen J, Tonini GP, Valteau-Couanet D, Varesio L, Versteeg R, Speleman F, Maris JM, Schleiermacher G, Preter KD | Genomic amplifications and distal 6q loss: novel markers for poor survival in high-risk neuroblastoma patients | JNCI: Journal of the National Cancer Institute 110(10) | Publisher |
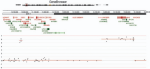 |
2018 | Depuydt P, Koster J, Boeva V, Hocking TD, Speleman F, Schleiermacher G, De Preter K | Meta-mining of copy number profiles of high-risk neuroblastoma tumors | Scientific data 5(1) | Publisher |
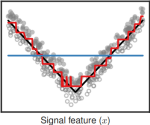 |
2017 | Drouin A, Hocking T, Laviolette F | Maximum Margin Interval Trees | Advances in Neural Information Processing Systems 30 | Publisher, Software, Reproducible, Preprint, Video |
 |
2017 | Hocking TD, Goerner-Potvin P, Morin A, Shao X, Pastinen T, Bourque G | Optimizing ChIP-seq peak detectors using visual labels and supervised machine learning | Bioinformatics 33(4) | Pubmed, Software, Reproducible, Data |
 |
2017 | Maidstone R, Hocking T, Rigaill G, Fearnhead P | On optimal multiple changepoint algorithms for large data | Statistics and Computing 27 | Publisher, Software, Reproducible |
 |
2016 | Chicard M, Boyault S, Colmet Daage L, Richer W, Gentien D, Pierron G, Lapouble E, Bellini A, Clement N, Iacono I, Bréjon Sé, Carrere M, Reyes Cé, Hocking T, Bernard V, Peuchmaur M, Corradini Nè, Faure-Conter Cé, Coze C, Plantaz D, Defachelles AS, Thebaud E, Gambart M, Millot Féé, Valteau-Couanet D, Michon J, Puisieux A, Delattre O, Combaret Vé, Schleiermacher G | Genomic Copy Number Profiling Using Circulating Free Tumor DNA Highlights Heterogeneity in Neuroblastoma | Clinical Cancer Research 22(22) | Publisher |
 |
2016 | Shimada K, Shimada S, Sugimoto K, Nakatochi M, Suguro M, Hirakawa A, Hocking TD, Takeuchi I, Tokunaga T, Takagi Y, Sakamoto A, Aoki T, Naoe T, Nakamura S, Hayakawa F, Seto M, Tomita A, Kiyoi H | Development and analysis of patient-derived xenograft mouse models in intravascular large B-cell lymphoma | Leukemia 30(7) | Pubmed |
 |
2015 | Hocking TD, Rigaill G, Bourque G | PeakSeg: constrained optimal segmentation and supervised penalty learning for peak detection in count data | Proc. 32nd ICML | Publisher, Video, Software, Reproducible |
 |
2014 | Hocking TD, Boeva V, Rigaill G, Schleiermacher G, Janoueix-Lerosey I, Delattre O, Richer W, Bourdeaut F, Suguro M, Seto M, Bach F, Vert JP | SegAnnDB: interactive Web-based genomic segmentation | Bioinformatics 30(11) | Pubmed, Software, Reproducible, Preprint, Package, Reproducible |
 |
2014 | Suguro M, Yoshida N, Umino A, Kato H, Tagawa H, Nakagawa M, Fukuhara N, Karnan S, Takeuchi I, Hocking TD, Arita K, Karube K, Tsuzuki S, Nakamura S, Kinoshita T, Seto M | Clonal heterogeneity of lymphoid malignancies correlates with poor prognosis | Cancer Sci 105(7) | Pubmed |
 |
2013 | Hocking TD, Schleiermacher G, Janoueix-Lerosey I, Boeva V, Cappo J, Delattre O, Bach F, Vert J-P | Learning smoothing models of copy number profiles using breakpoint annotations | BMC Bioinformatics 14(164) | Publisher, Software, Reproducible |
 |
2013 | Hocking TD, Wutzler T, Ponting K, Grosjean P | Sustainable, Extensible Documentation Generation using inlinedocs | Journal of Statistical Software 54 | Publisher, Software, Reproducible |
 |
2013 | Rigaill G, Hocking T, Vert J-P, Bach F | Learning sparse penalties for change-point detection using max margin interval regression | Proc. 30th ICML | Publisher, Video, Software, Reproducible |
 |
2012 | Hocking TD | Learning algorithms and statistical software, with applications to bioinformatics | PHD thesis, Ecole normale supérieure de Cachan | Publisher, Reproducible |
 |
2011 | Hocking TD, Joulin A, Bach F, Vert J-P | Clusterpath an algorithm for clustering using convex fusion penalties | 28th international conference on machine learning | Publisher, Video, Software, Reproducible, Cited in Ihaka lecture |
 |
2010 | Gautier M, Hocking TD, Foulley J-L | A Bayesian outlier criterion to detect SNPs under selection in large data sets | PLoS one 5(8) | Publisher |
 |
2008 | Doyon Y, McCammon JM, Miller JC, Faraji F, Ngo C, Katibah GE, Amora R, Hocking TD, Zhang L, Rebar EJ, Gregory PD, Urnov FD, Amacher SL | Heritable targeted gene disruption in zebrafish using designed zinc-finger nucleases | Nature biotechnology 26(6) | Pubmed |
The output above is a table with one row per publication, and an image column that shows a figure from the paper. The trick to getting that to display, is putting it in this repo, with a standard name, based on the bib file key.
The code below checks for missing figures.
some.out[Figure=='', Title]
## character(0)
Make sure pdflatex likes it
This part only works in Rmd, not md/jekyll for some reason.
Report mis-match between image files and refs
img.dt <- data.table(ref=sub(".png", "", dir("../assets/img/publications/")))
img.dt[!refs.wide,on="ref"] #images without bib entries
## ref
## <char>
## 1: thumb
refs.wide[!img.dt,.(ref),on="ref"] #bib entries without images
## Empty data.table (0 rows and 1 cols): ref
Conclusion
We have seen how a bib file can be used to define a publications web page.
- bib file contains peer-reviewed, published papers as
article(for journals) andinproceedings(for conferences), incollectionis used for book chapters,phdthesisis used for PHD thesis,
unpublished is used for papers in progress (not yet published), meaning one or more of below:
- conference tutorial, note should include which conference; links should include conference page, book/manual page, and github source.
- paper not yet submitted for review (note=In progress),
- Pre-print available, put in note and links.
- If under review or accepted, put venue in note.
Because bibtex ignores fields like links which are not part of
standard bibliography types, we can put markdown code in there, and
then put it into our markdown pubs page.
Session info
sessionInfo()
## R version 4.5.2 (2025-10-31 ucrt)
## Platform: x86_64-w64-mingw32/x64
## Running under: Windows 11 x64 (build 26100)
##
## Matrix products: default
## LAPACK version 3.12.1
##
## locale:
## [1] LC_COLLATE=English_United States.utf8 LC_CTYPE=English_United States.utf8 LC_MONETARY=English_United States.utf8
## [4] LC_NUMERIC=C LC_TIME=English_United States.utf8
##
## time zone: America/Toronto
## tzcode source: internal
##
## attached base packages:
## [1] stats graphics utils datasets grDevices methods base
##
## other attached packages:
## [1] data.table_1.17.99
##
## loaded via a namespace (and not attached):
## [1] compiler_4.5.2 nc_2025.3.24 tools_4.5.2 knitr_1.50 xfun_0.54 evaluate_1.0.5