data.table asymptotic timings
The purpose of this vignette is to make figures which show the efficiency of data.table.
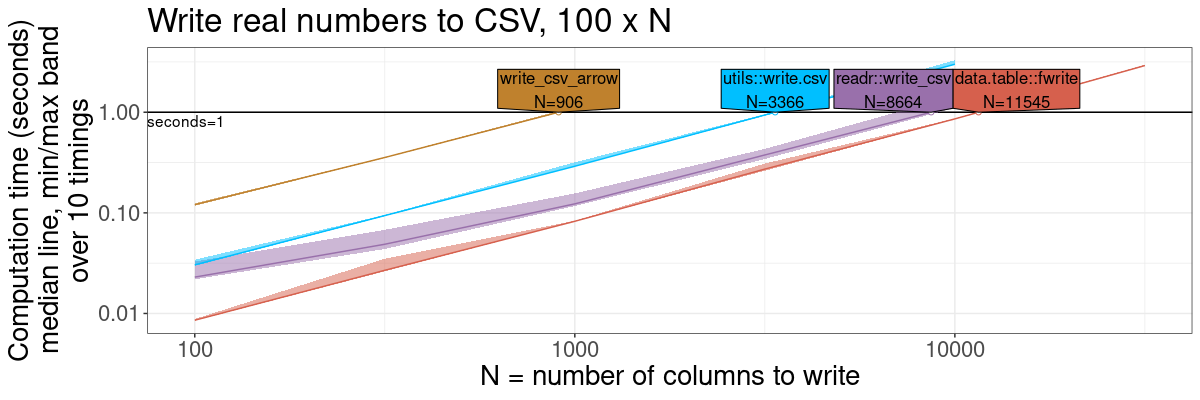
fwrite: fast CSV writer
library(data.table)
library(readr)
library(arrow)
library(ggplot2)
write.colors <- c(
"readr::write_csv"="#9970AB",
"data.table::fwrite"="#D6604D",
"write_csv_arrow"="#BF812D",
"utils::write.csv"="deepskyblue")
n.rows <- 100
seconds.limit <- 1
atime.write.vary.cols <- atime::atime(
N=as.integer(10^seq(2, 6, by=0.5)),
setup={
set.seed(1)
input.vec <- rnorm(n.rows*N)
input.mat <- matrix(input.vec, n.rows, N)
input.df <- data.frame(input.mat)
},
seconds.limit = seconds.limit,
"data.table::fwrite"={
data.table::fwrite(input.df, tempfile(), showProgress = FALSE)
},
"write_csv_arrow"={
arrow::write_csv_arrow(input.df, tempfile())
},
"readr::write_csv"={
readr::write_csv(input.df, tempfile(), progress = FALSE)
},
"utils::write.csv"=utils::write.csv(input.df, tempfile()))
## Warning: Some expressions had a GC in every iteration; so filtering is disabled.
refs.write.vary.cols <- atime::references_best(atime.write.vary.cols)
pred.write.vary.cols <- predict(refs.write.vary.cols)
gg.write <- plot(pred.write.vary.cols)+
theme(text=element_text(size=20))+
ggtitle(sprintf("Write real numbers to CSV, %d x N", n.rows))+
scale_x_log10("N = number of columns to write")+
scale_y_log10("Computation time (seconds)
median line, min/max band
over 10 timings")+
facet_null()+
scale_fill_manual(values=write.colors)+
scale_color_manual(values=write.colors)
## Scale for x is already present.
## Adding another scale for x, which will replace the existing scale.
## Scale for y is already present.
## Adding another scale for y, which will replace the existing scale.
gg.write
## Warning: Transformation introduced infinite values in continuous x-axis
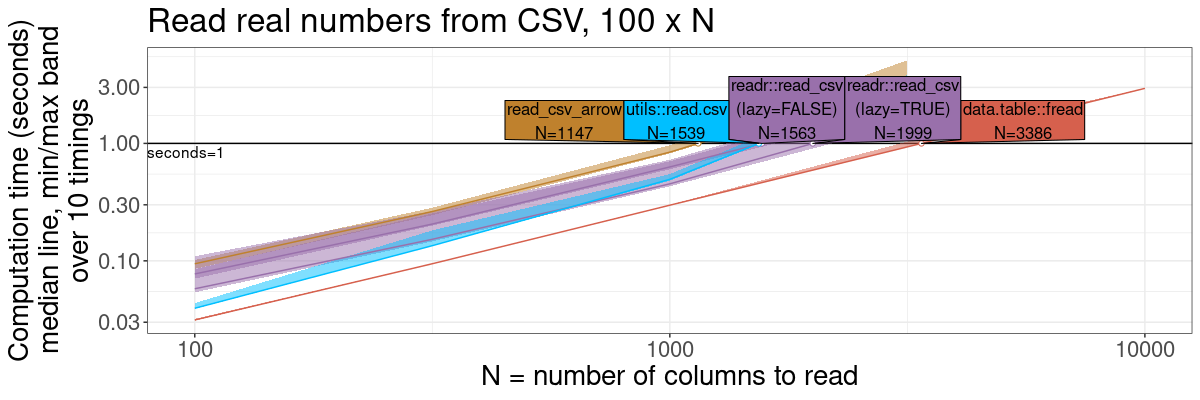
fread: fast CSV reader
read.colors <- c(
"readr::read_csv\n(lazy=TRUE)"="#9970AB",
"readr::read_csv\n(lazy=FALSE)"="#9970AB",
"data.table::fread"="#D6604D",
"read_csv_arrow"="#BF812D",
"utils::read.csv"="deepskyblue")
atime.read.vary.cols <- atime::atime(
N=as.integer(10^seq(2, 6, by=0.5)),
setup={
set.seed(1)
input.vec <- rnorm(n.rows*N)
input.mat <- matrix(input.vec, n.rows, N)
input.df <- data.frame(input.mat)
input.csv <- tempfile()
fwrite(input.df, input.csv)
},
seconds.limit = seconds.limit,
"data.table::fread"={
data.table::fread(input.csv, showProgress = FALSE)
},
"read_csv_arrow"={
arrow::read_csv_arrow(input.csv)
},
"readr::read_csv\n(lazy=TRUE)"={
readr::read_csv(input.csv, progress = FALSE, show_col_types = FALSE, lazy=TRUE)
},
"readr::read_csv\n(lazy=FALSE)"={
readr::read_csv(input.csv, progress = FALSE, show_col_types = FALSE, lazy=FALSE)
},
"utils::read.csv"=utils::read.csv(input.csv))
## Warning: Some expressions had a GC in every iteration; so filtering is disabled.
refs.read.vary.cols <- atime::references_best(atime.read.vary.cols)
pred.read.vary.cols <- predict(refs.read.vary.cols)
gg.read <- plot(pred.read.vary.cols)+
theme(text=element_text(size=20))+
ggtitle(sprintf("Read real numbers from CSV, %d x N", n.rows))+
scale_x_log10("N = number of columns to read")+
scale_y_log10("Computation time (seconds)
median line, min/max band
over 10 timings")+
facet_null()+
scale_fill_manual(values=read.colors)+
scale_color_manual(values=read.colors)
## Scale for x is already present.
## Adding another scale for x, which will replace the existing scale.
## Scale for y is already present.
## Adding another scale for y, which will replace the existing scale.
gg.read
## Warning: Transformation introduced infinite values in continuous x-axis
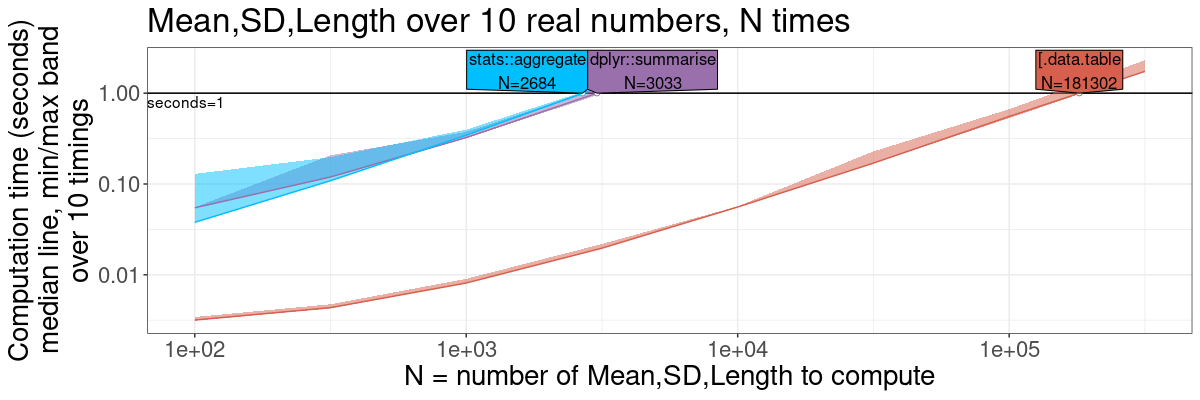
Summarize by group
ml.colors <- c(
"dplyr::summarise"="#9970AB",
"[.data.table"="#D6604D",
"stats::aggregate"="deepskyblue")
options(dplyr.summarise.inform=FALSE)
n.folds <- 10
ml.atime <- atime::atime(
N=as.integer(10^seq(2, 7, by=0.5)),
setup={
loss.dt <- data.table(
name="loss",
fold=rep(1:n.folds, each=2*N),
loss=rnorm(2*N*n.folds),
set=rep(c("subtrain","validation"),each=N),
epoch=1:N,
key=c("set","epoch","fold"))
},
seconds.limit=seconds.limit,
"[.data.table"={
loss.dt[, .(
loss_length=.N,
loss_mean=mean(loss),
loss_sd=sd(loss)
), by=.(set, epoch)]
},
"stats::aggregate"={
res <- stats::aggregate(
loss ~ set + epoch,
loss.dt,
function(values)list(c(
loss_length=length(values),
loss_mean=mean(values),
loss_sd=sd(values))))
data.frame(
subset(res, select=-loss),
do.call(rbind, res$loss))
},
"dplyr::summarise"={
loss.dt |>
dplyr::group_by(set, epoch) |>
dplyr::summarise(
loss_length=length(loss),
loss_mean=mean(loss),
loss_sd=sd(loss))
})
## Warning: Some expressions had a GC in every iteration; so filtering is disabled.
ml.refs <- atime::references_best(ml.atime)
ml.pred <- predict(ml.refs)
ml.gg <- plot(ml.pred)+
theme(text=element_text(size=20))+
ggtitle(sprintf("Mean,SD,Length over %d real numbers, N times", n.folds))+
scale_x_log10("N = number of Mean,SD,Length to compute")+
scale_y_log10("Computation time (seconds)
median line, min/max band
over 10 timings")+
facet_null()+
scale_fill_manual(values=ml.colors)+
scale_color_manual(values=ml.colors)
## Scale for x is already present.
## Adding another scale for x, which will replace the existing scale.
## Scale for y is already present.
## Adding another scale for y, which will replace the existing scale.
ml.gg
## Warning: Transformation introduced infinite values in continuous x-axis
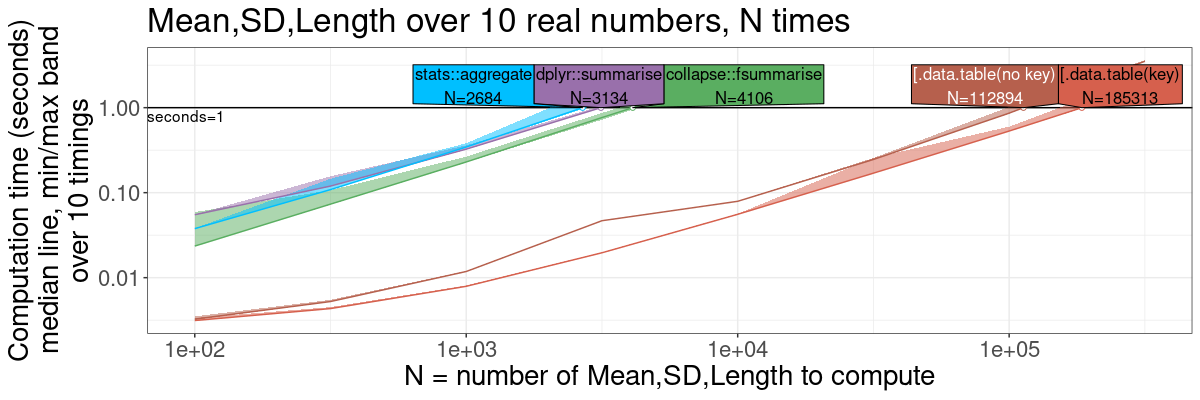
Summarize by group, expanded
The previous section is simpler to explain, whereas this section is
more comprehensive/fair, because it shows versions of data.table
with and without key. Both versions are much faster than the
competitors, though.
options(dplyr.summarise.inform=FALSE)
n.folds <- 10
ml.exp.atime <- atime::atime(
N=as.integer(10^seq(2, 7, by=0.5)),
setup={
loss.dt <- data.table(
name="loss",
fold=rep(1:n.folds, each=2*N),
loss=rnorm(2*N*n.folds),
set=rep(c("subtrain","validation"),each=N),
epoch=1:N)
key.dt <- data.table(loss.dt, key=c("set","epoch","fold"))
},
seconds.limit=seconds.limit,
"[.data.table(no key)"={
loss.dt[, .(
loss_length=.N,
loss_mean=mean(loss),
loss_sd=sd(loss)
), by=.(set, epoch)]
},
"[.data.table(key)"={
key.dt[, .(
loss_length=.N,
loss_mean=mean(loss),
loss_sd=sd(loss)
), by=.(set, epoch)]
},
"stats::aggregate"={
res <- stats::aggregate(
loss ~ set + epoch,
loss.dt,
function(values)list(c(
loss_length=length(values),
loss_mean=mean(values),
loss_sd=sd(values))))
data.frame(
subset(res, select=-loss),
do.call(rbind, res$loss))
},
"dplyr::summarise"={
loss.dt |>
dplyr::group_by(set, epoch) |>
dplyr::summarise(
loss_length=length(loss),
loss_mean=mean(loss),
loss_sd=sd(loss))
},
"collapse::fsummarise"={
loss.dt |>
collapse::fgroup_by(set, epoch) |>
collapse::fsummarise(
loss_length=length(loss),
loss_mean=mean(loss),
loss_sd=sd(loss))
})
## Warning: Some expressions had a GC in every iteration; so filtering is disabled.
## Warning: Some expressions had a GC in every iteration; so filtering is disabled.
## Warning: Some expressions had a GC in every iteration; so filtering is disabled.
ml.exp.refs <- atime::references_best(ml.exp.atime)
ml.exp.pred <- predict(ml.exp.refs)
ml.exp.colors <- c(
"collapse::fsummarise"="#5AAE61",
"dplyr::summarise"="#9970AB",
"[.data.table(key)"="#D6604D",
"[.data.table(no key)"="#B6604D",
"stats::aggregate"="deepskyblue")
ml.exp.gg <- plot(ml.exp.pred)+
theme(text=element_text(size=20))+
ggtitle(sprintf("Mean,SD,Length over %d real numbers, N times", n.folds))+
scale_x_log10("N = number of Mean,SD,Length to compute")+
scale_y_log10("Computation time (seconds)
median line, min/max band
over 10 timings")+
facet_null()+
scale_fill_manual(values=ml.exp.colors)+
scale_color_manual(values=ml.exp.colors)
## Scale for x is already present.
## Adding another scale for x, which will replace the existing scale.
## Scale for y is already present.
## Adding another scale for y, which will replace the existing scale.
ml.exp.gg
## Warning: Transformation introduced infinite values in continuous x-axis
version info
sessionInfo()
## R version 4.3.1 (2023-06-16)
## Platform: x86_64-pc-linux-gnu (64-bit)
## Running under: Ubuntu 22.04.3 LTS
##
## Matrix products: default
## BLAS: /usr/lib/x86_64-linux-gnu/blas/libblas.so.3.10.0
## LAPACK: /usr/lib/x86_64-linux-gnu/lapack/liblapack.so.3.10.0
##
## locale:
## [1] LC_CTYPE=fr_FR.UTF-8 LC_NUMERIC=C LC_TIME=fr_FR.UTF-8
## [4] LC_COLLATE=fr_FR.UTF-8 LC_MONETARY=fr_FR.UTF-8 LC_MESSAGES=fr_FR.UTF-8
## [7] LC_PAPER=fr_FR.UTF-8 LC_NAME=C LC_ADDRESS=C
## [10] LC_TELEPHONE=C LC_MEASUREMENT=fr_FR.UTF-8 LC_IDENTIFICATION=C
##
## time zone: America/Phoenix
## tzcode source: system (glibc)
##
## attached base packages:
## [1] stats graphics utils datasets grDevices methods base
##
## other attached packages:
## [1] ggplot2_3.4.3 arrow_13.0.0 readr_2.1.4 data.table_1.14.8
##
## loaded via a namespace (and not attached):
## [1] bit_4.0.5 gtable_0.3.4 highr_0.10 dplyr_1.1.2
## [5] compiler_4.3.1 crayon_1.5.2 Rcpp_1.0.11 tidyselect_1.2.0
## [9] collapse_1.9.6 parallel_4.3.1 assertthat_0.2.1 scales_1.2.1
## [13] directlabels_2023.8.25 lattice_0.21-8 R6_2.5.1 generics_0.1.3
## [17] knitr_1.43 tibble_3.2.1 munsell_0.5.0 atime_2023.10.9
## [21] pillar_1.9.0 tzdb_0.3.0 rlang_1.1.1 utf8_1.2.3
## [25] xfun_0.40 quadprog_1.5-8 bit64_4.0.5 cli_3.6.1
## [29] withr_2.5.0 magrittr_2.0.3 grid_4.3.1 vroom_1.6.3
## [33] hms_1.1.3 lifecycle_1.0.3 vctrs_0.6.3 bench_1.1.3
## [37] evaluate_0.21 glue_1.6.2 farver_2.1.1 profmem_0.6.0
## [41] fansi_1.0.4 colorspace_2.1-0 purrr_1.0.2 tools_4.3.1
## [45] pkgconfig_2.0.3