R Package Release History
For a recent grant proposal submission to the National Science
Foundation POSE program, I wanted to make a figure that shows the
release history of the data.table R package.
Download Archive web page
First, we create a releases directory to cache the downloaded web pages,
releases.dir <- "~/grants/2022-NSF-POSE/releases"
dir.create(releases.dir, showWarnings = FALSE)
We can download an Archive web page via the code below,
Archive <- "https://cloud.r-project.org/src/contrib/Archive/"
get_Archive <- function(Package){
pkg.html <- file.path(releases.dir, paste0(Package, ".html"))
if(!file.exists(pkg.html)){
u <- paste0(Archive, Package)
download.file(u, pkg.html)
}
readLines(pkg.html)
}
(Archive.data.table <- get_Archive("data.table"))
## [1] "<!DOCTYPE HTML PUBLIC \"-//W3C//DTD HTML 3.2 Final//EN\">"
## [2] "<html>"
## [3] " <head>"
## [4] " <title>Index of /src/contrib/Archive/data.table</title>"
## [5] " </head>"
## [6] " <body>"
## [7] "<h1>Index of /src/contrib/Archive/data.table</h1>"
## [8] "<pre> <a href=\"?C=N;O=D\">Name</a> <a href=\"?C=M;O=A\">Last modified</a> <a href=\"?C=S;O=A\">Size</a> <hr> <a href=\"/src/contrib/Archive/\">Parent Directory</a> - "
## [9] " <a href=\"data.table_1.0.tar.gz\">data.table_1.0.tar.gz</a> 2006-04-14 22:03 16K "
## [10] " <a href=\"data.table_1.1.tar.gz\">data.table_1.1.tar.gz</a> 2008-08-27 07:35 40K "
## [11] " <a href=\"data.table_1.10.0.tar.gz\">data.table_1.10.0.tar.gz</a> 2016-12-03 10:05 2.9M "
## [12] " <a href=\"data.table_1.10.2.tar.gz\">data.table_1.10.2.tar.gz</a> 2017-01-31 15:09 2.9M "
## [13] " <a href=\"data.table_1.10.4-1.tar.gz\">data.table_1.10.4-1.tar.gz</a> 2017-10-09 22:36 2.9M "
## [14] " <a href=\"data.table_1.10.4-2.tar.gz\">data.table_1.10.4-2.tar.gz</a> 2017-10-12 14:03 2.9M "
## [15] " <a href=\"data.table_1.10.4-3.tar.gz\">data.table_1.10.4-3.tar.gz</a> 2017-10-27 07:40 2.9M "
## [16] " <a href=\"data.table_1.10.4.tar.gz\">data.table_1.10.4.tar.gz</a> 2017-02-01 14:52 2.9M "
## [17] " <a href=\"data.table_1.11.0.tar.gz\">data.table_1.11.0.tar.gz</a> 2018-05-01 17:00 3.1M "
## [18] " <a href=\"data.table_1.11.2.tar.gz\">data.table_1.11.2.tar.gz</a> 2018-05-08 16:16 3.1M "
## [19] " <a href=\"data.table_1.11.4.tar.gz\">data.table_1.11.4.tar.gz</a> 2018-05-27 16:34 3.1M "
## [20] " <a href=\"data.table_1.11.6.tar.gz\">data.table_1.11.6.tar.gz</a> 2018-09-19 22:10 3.2M "
## [21] " <a href=\"data.table_1.11.8.tar.gz\">data.table_1.11.8.tar.gz</a> 2018-09-30 13:30 3.1M "
## [22] " <a href=\"data.table_1.12.0.tar.gz\">data.table_1.12.0.tar.gz</a> 2019-01-13 11:50 3.2M "
## [23] " <a href=\"data.table_1.12.2.tar.gz\">data.table_1.12.2.tar.gz</a> 2019-04-07 10:06 3.2M "
## [24] " <a href=\"data.table_1.12.4.tar.gz\">data.table_1.12.4.tar.gz</a> 2019-10-03 09:10 4.8M "
## [25] " <a href=\"data.table_1.12.6.tar.gz\">data.table_1.12.6.tar.gz</a> 2019-10-18 22:20 4.7M "
## [26] " <a href=\"data.table_1.12.8.tar.gz\">data.table_1.12.8.tar.gz</a> 2019-12-09 10:30 4.7M "
## [27] " <a href=\"data.table_1.13.0.tar.gz\">data.table_1.13.0.tar.gz</a> 2020-07-24 09:40 5.0M "
## [28] " <a href=\"data.table_1.13.2.tar.gz\">data.table_1.13.2.tar.gz</a> 2020-10-19 18:50 5.0M "
## [29] " <a href=\"data.table_1.13.4.tar.gz\">data.table_1.13.4.tar.gz</a> 2020-12-08 10:10 5.0M "
## [30] " <a href=\"data.table_1.13.6.tar.gz\">data.table_1.13.6.tar.gz</a> 2020-12-30 15:50 5.1M "
## [31] " <a href=\"data.table_1.14.0.tar.gz\">data.table_1.14.0.tar.gz</a> 2021-02-21 06:00 5.1M "
## [32] " <a href=\"data.table_1.2.tar.gz\">data.table_1.2.tar.gz</a> 2008-09-01 06:59 40K "
## [33] " <a href=\"data.table_1.4.1.tar.gz\">data.table_1.4.1.tar.gz</a> 2010-05-03 08:40 344K "
## [34] " <a href=\"data.table_1.5.1.tar.gz\">data.table_1.5.1.tar.gz</a> 2011-01-08 08:31 589K "
## [35] " <a href=\"data.table_1.5.2.tar.gz\">data.table_1.5.2.tar.gz</a> 2011-01-21 09:03 607K "
## [36] " <a href=\"data.table_1.5.3.tar.gz\">data.table_1.5.3.tar.gz</a> 2011-02-11 08:49 623K "
## [37] " <a href=\"data.table_1.5.tar.gz\">data.table_1.5.tar.gz</a> 2010-09-14 06:23 589K "
## [38] " <a href=\"data.table_1.6.1.tar.gz\">data.table_1.6.1.tar.gz</a> 2011-06-29 09:41 692K "
## [39] " <a href=\"data.table_1.6.2.tar.gz\">data.table_1.6.2.tar.gz</a> 2011-07-02 14:21 693K "
## [40] " <a href=\"data.table_1.6.3.tar.gz\">data.table_1.6.3.tar.gz</a> 2011-08-04 11:28 698K "
## [41] " <a href=\"data.table_1.6.4.tar.gz\">data.table_1.6.4.tar.gz</a> 2011-08-10 05:50 705K "
## [42] " <a href=\"data.table_1.6.5.tar.gz\">data.table_1.6.5.tar.gz</a> 2011-08-25 04:35 711K "
## [43] " <a href=\"data.table_1.6.6.tar.gz\">data.table_1.6.6.tar.gz</a> 2011-08-25 20:08 712K "
## [44] " <a href=\"data.table_1.6.tar.gz\">data.table_1.6.tar.gz</a> 2011-04-24 06:07 684K "
## [45] " <a href=\"data.table_1.7.1.tar.gz\">data.table_1.7.1.tar.gz</a> 2011-10-22 12:05 728K "
## [46] " <a href=\"data.table_1.7.10.tar.gz\">data.table_1.7.10.tar.gz</a> 2012-02-07 08:43 758K "
## [47] " <a href=\"data.table_1.7.2.tar.gz\">data.table_1.7.2.tar.gz</a> 2011-11-07 14:05 735K "
## [48] " <a href=\"data.table_1.7.3.tar.gz\">data.table_1.7.3.tar.gz</a> 2011-11-25 07:12 741K "
## [49] " <a href=\"data.table_1.7.4.tar.gz\">data.table_1.7.4.tar.gz</a> 2011-11-29 06:57 741K "
## [50] " <a href=\"data.table_1.7.5.tar.gz\">data.table_1.7.5.tar.gz</a> 2011-12-04 12:51 742K "
## [51] " <a href=\"data.table_1.7.6.tar.gz\">data.table_1.7.6.tar.gz</a> 2011-12-13 08:36 743K "
## [52] " <a href=\"data.table_1.7.7.tar.gz\">data.table_1.7.7.tar.gz</a> 2011-12-15 10:07 744K "
## [53] " <a href=\"data.table_1.7.8.tar.gz\">data.table_1.7.8.tar.gz</a> 2012-01-25 07:53 754K "
## [54] " <a href=\"data.table_1.7.9.tar.gz\">data.table_1.7.9.tar.gz</a> 2012-01-31 07:30 756K "
## [55] " <a href=\"data.table_1.8.0.tar.gz\">data.table_1.8.0.tar.gz</a> 2012-07-16 08:21 768K "
## [56] " <a href=\"data.table_1.8.10.tar.gz\">data.table_1.8.10.tar.gz</a> 2013-09-03 04:41 914K "
## [57] " <a href=\"data.table_1.8.2.tar.gz\">data.table_1.8.2.tar.gz</a> 2012-07-17 19:51 799K "
## [58] " <a href=\"data.table_1.8.4.tar.gz\">data.table_1.8.4.tar.gz</a> 2012-11-09 15:23 820K "
## [59] " <a href=\"data.table_1.8.6.tar.gz\">data.table_1.8.6.tar.gz</a> 2012-11-13 13:28 821K "
## [60] " <a href=\"data.table_1.8.8.tar.gz\">data.table_1.8.8.tar.gz</a> 2013-03-06 06:31 874K "
## [61] " <a href=\"data.table_1.9.2.tar.gz\">data.table_1.9.2.tar.gz</a> 2014-02-27 13:49 1.0M "
## [62] " <a href=\"data.table_1.9.4.tar.gz\">data.table_1.9.4.tar.gz</a> 2014-10-02 06:41 927K "
## [63] " <a href=\"data.table_1.9.6.tar.gz\">data.table_1.9.6.tar.gz</a> 2015-09-19 20:13 3.5M "
## [64] " <a href=\"data.table_1.9.8.tar.gz\">data.table_1.9.8.tar.gz</a> 2016-11-25 11:55 2.9M "
## [65] "<hr></pre>"
## [66] "<address>Apache/2.4.39 (Unix) Server at cloud.r-project.org Port 80</address>"
## [67] "</body></html>"
The output above shows that the Archive web page has a regular structure.
Parsing with a regular expression
The Archive web page can be parsed via a regular expression,
naive.pattern <- list(
"_",
version="[0-9.]+",
"[.]tar[.]gz</a>\\s+",
date.str=".*?",
"\\s")
The code above specifies a regular expression:
"_"means to start by matching an underscore,version="[0-9.]+"means to match one or more digits or dots, and output them in theversioncolumn,"[.]tar[.]gz</a>\\s+"means to match the.tar.gzfile name suffix, the closing</a>tag, and then one or more white space characters,date.str=".*?"means to match zero or more characters (non-greedy, as few as possible), and output them in thedate.strcolumn,"\\s"means to match one white space character.
The end result is a table with one row for each matched package version, and one column for each of the named arguments:
(naive.dt <- nc::capture_all_str(Archive.data.table, naive.pattern))
## version date.str
## <char> <char>
## 1: 1.0 2006-04-14
## 2: 1.1 2008-08-27
## 3: 1.10.0 2016-12-03
## 4: 1.10.2 2017-01-31
## 5: 1.10.4 2017-02-01
## 6: 1.11.0 2018-05-01
## 7: 1.11.2 2018-05-08
## 8: 1.11.4 2018-05-27
## 9: 1.11.6 2018-09-19
## 10: 1.11.8 2018-09-30
## 11: 1.12.0 2019-01-13
## 12: 1.12.2 2019-04-07
## 13: 1.12.4 2019-10-03
## 14: 1.12.6 2019-10-18
## 15: 1.12.8 2019-12-09
## 16: 1.13.0 2020-07-24
## 17: 1.13.2 2020-10-19
## 18: 1.13.4 2020-12-08
## 19: 1.13.6 2020-12-30
## 20: 1.14.0 2021-02-21
## 21: 1.2 2008-09-01
## 22: 1.4.1 2010-05-03
## 23: 1.5.1 2011-01-08
## 24: 1.5.2 2011-01-21
## 25: 1.5.3 2011-02-11
## 26: 1.5 2010-09-14
## 27: 1.6.1 2011-06-29
## 28: 1.6.2 2011-07-02
## 29: 1.6.3 2011-08-04
## 30: 1.6.4 2011-08-10
## 31: 1.6.5 2011-08-25
## 32: 1.6.6 2011-08-25
## 33: 1.6 2011-04-24
## 34: 1.7.1 2011-10-22
## 35: 1.7.10 2012-02-07
## 36: 1.7.2 2011-11-07
## 37: 1.7.3 2011-11-25
## 38: 1.7.4 2011-11-29
## 39: 1.7.5 2011-12-04
## 40: 1.7.6 2011-12-13
## 41: 1.7.7 2011-12-15
## 42: 1.7.8 2012-01-25
## 43: 1.7.9 2012-01-31
## 44: 1.8.0 2012-07-16
## 45: 1.8.10 2013-09-03
## 46: 1.8.2 2012-07-17
## 47: 1.8.4 2012-11-09
## 48: 1.8.6 2012-11-13
## 49: 1.8.8 2013-03-06
## 50: 1.9.2 2014-02-27
## 51: 1.9.4 2014-10-02
## 52: 1.9.6 2015-09-19
## 53: 1.9.8 2016-11-25
## version date.str
Above the table shows all matches for the given pattern. How do we know that it has captured all the patterns? We can use a simpler pattern to get all of the packages,
(tar.gz.vec <- grep("tar[.]gz", Archive.data.table, value=TRUE))
## [1] " <a href=\"data.table_1.0.tar.gz\">data.table_1.0.tar.gz</a> 2006-04-14 22:03 16K "
## [2] " <a href=\"data.table_1.1.tar.gz\">data.table_1.1.tar.gz</a> 2008-08-27 07:35 40K "
## [3] " <a href=\"data.table_1.10.0.tar.gz\">data.table_1.10.0.tar.gz</a> 2016-12-03 10:05 2.9M "
## [4] " <a href=\"data.table_1.10.2.tar.gz\">data.table_1.10.2.tar.gz</a> 2017-01-31 15:09 2.9M "
## [5] " <a href=\"data.table_1.10.4-1.tar.gz\">data.table_1.10.4-1.tar.gz</a> 2017-10-09 22:36 2.9M "
## [6] " <a href=\"data.table_1.10.4-2.tar.gz\">data.table_1.10.4-2.tar.gz</a> 2017-10-12 14:03 2.9M "
## [7] " <a href=\"data.table_1.10.4-3.tar.gz\">data.table_1.10.4-3.tar.gz</a> 2017-10-27 07:40 2.9M "
## [8] " <a href=\"data.table_1.10.4.tar.gz\">data.table_1.10.4.tar.gz</a> 2017-02-01 14:52 2.9M "
## [9] " <a href=\"data.table_1.11.0.tar.gz\">data.table_1.11.0.tar.gz</a> 2018-05-01 17:00 3.1M "
## [10] " <a href=\"data.table_1.11.2.tar.gz\">data.table_1.11.2.tar.gz</a> 2018-05-08 16:16 3.1M "
## [11] " <a href=\"data.table_1.11.4.tar.gz\">data.table_1.11.4.tar.gz</a> 2018-05-27 16:34 3.1M "
## [12] " <a href=\"data.table_1.11.6.tar.gz\">data.table_1.11.6.tar.gz</a> 2018-09-19 22:10 3.2M "
## [13] " <a href=\"data.table_1.11.8.tar.gz\">data.table_1.11.8.tar.gz</a> 2018-09-30 13:30 3.1M "
## [14] " <a href=\"data.table_1.12.0.tar.gz\">data.table_1.12.0.tar.gz</a> 2019-01-13 11:50 3.2M "
## [15] " <a href=\"data.table_1.12.2.tar.gz\">data.table_1.12.2.tar.gz</a> 2019-04-07 10:06 3.2M "
## [16] " <a href=\"data.table_1.12.4.tar.gz\">data.table_1.12.4.tar.gz</a> 2019-10-03 09:10 4.8M "
## [17] " <a href=\"data.table_1.12.6.tar.gz\">data.table_1.12.6.tar.gz</a> 2019-10-18 22:20 4.7M "
## [18] " <a href=\"data.table_1.12.8.tar.gz\">data.table_1.12.8.tar.gz</a> 2019-12-09 10:30 4.7M "
## [19] " <a href=\"data.table_1.13.0.tar.gz\">data.table_1.13.0.tar.gz</a> 2020-07-24 09:40 5.0M "
## [20] " <a href=\"data.table_1.13.2.tar.gz\">data.table_1.13.2.tar.gz</a> 2020-10-19 18:50 5.0M "
## [21] " <a href=\"data.table_1.13.4.tar.gz\">data.table_1.13.4.tar.gz</a> 2020-12-08 10:10 5.0M "
## [22] " <a href=\"data.table_1.13.6.tar.gz\">data.table_1.13.6.tar.gz</a> 2020-12-30 15:50 5.1M "
## [23] " <a href=\"data.table_1.14.0.tar.gz\">data.table_1.14.0.tar.gz</a> 2021-02-21 06:00 5.1M "
## [24] " <a href=\"data.table_1.2.tar.gz\">data.table_1.2.tar.gz</a> 2008-09-01 06:59 40K "
## [25] " <a href=\"data.table_1.4.1.tar.gz\">data.table_1.4.1.tar.gz</a> 2010-05-03 08:40 344K "
## [26] " <a href=\"data.table_1.5.1.tar.gz\">data.table_1.5.1.tar.gz</a> 2011-01-08 08:31 589K "
## [27] " <a href=\"data.table_1.5.2.tar.gz\">data.table_1.5.2.tar.gz</a> 2011-01-21 09:03 607K "
## [28] " <a href=\"data.table_1.5.3.tar.gz\">data.table_1.5.3.tar.gz</a> 2011-02-11 08:49 623K "
## [29] " <a href=\"data.table_1.5.tar.gz\">data.table_1.5.tar.gz</a> 2010-09-14 06:23 589K "
## [30] " <a href=\"data.table_1.6.1.tar.gz\">data.table_1.6.1.tar.gz</a> 2011-06-29 09:41 692K "
## [31] " <a href=\"data.table_1.6.2.tar.gz\">data.table_1.6.2.tar.gz</a> 2011-07-02 14:21 693K "
## [32] " <a href=\"data.table_1.6.3.tar.gz\">data.table_1.6.3.tar.gz</a> 2011-08-04 11:28 698K "
## [33] " <a href=\"data.table_1.6.4.tar.gz\">data.table_1.6.4.tar.gz</a> 2011-08-10 05:50 705K "
## [34] " <a href=\"data.table_1.6.5.tar.gz\">data.table_1.6.5.tar.gz</a> 2011-08-25 04:35 711K "
## [35] " <a href=\"data.table_1.6.6.tar.gz\">data.table_1.6.6.tar.gz</a> 2011-08-25 20:08 712K "
## [36] " <a href=\"data.table_1.6.tar.gz\">data.table_1.6.tar.gz</a> 2011-04-24 06:07 684K "
## [37] " <a href=\"data.table_1.7.1.tar.gz\">data.table_1.7.1.tar.gz</a> 2011-10-22 12:05 728K "
## [38] " <a href=\"data.table_1.7.10.tar.gz\">data.table_1.7.10.tar.gz</a> 2012-02-07 08:43 758K "
## [39] " <a href=\"data.table_1.7.2.tar.gz\">data.table_1.7.2.tar.gz</a> 2011-11-07 14:05 735K "
## [40] " <a href=\"data.table_1.7.3.tar.gz\">data.table_1.7.3.tar.gz</a> 2011-11-25 07:12 741K "
## [41] " <a href=\"data.table_1.7.4.tar.gz\">data.table_1.7.4.tar.gz</a> 2011-11-29 06:57 741K "
## [42] " <a href=\"data.table_1.7.5.tar.gz\">data.table_1.7.5.tar.gz</a> 2011-12-04 12:51 742K "
## [43] " <a href=\"data.table_1.7.6.tar.gz\">data.table_1.7.6.tar.gz</a> 2011-12-13 08:36 743K "
## [44] " <a href=\"data.table_1.7.7.tar.gz\">data.table_1.7.7.tar.gz</a> 2011-12-15 10:07 744K "
## [45] " <a href=\"data.table_1.7.8.tar.gz\">data.table_1.7.8.tar.gz</a> 2012-01-25 07:53 754K "
## [46] " <a href=\"data.table_1.7.9.tar.gz\">data.table_1.7.9.tar.gz</a> 2012-01-31 07:30 756K "
## [47] " <a href=\"data.table_1.8.0.tar.gz\">data.table_1.8.0.tar.gz</a> 2012-07-16 08:21 768K "
## [48] " <a href=\"data.table_1.8.10.tar.gz\">data.table_1.8.10.tar.gz</a> 2013-09-03 04:41 914K "
## [49] " <a href=\"data.table_1.8.2.tar.gz\">data.table_1.8.2.tar.gz</a> 2012-07-17 19:51 799K "
## [50] " <a href=\"data.table_1.8.4.tar.gz\">data.table_1.8.4.tar.gz</a> 2012-11-09 15:23 820K "
## [51] " <a href=\"data.table_1.8.6.tar.gz\">data.table_1.8.6.tar.gz</a> 2012-11-13 13:28 821K "
## [52] " <a href=\"data.table_1.8.8.tar.gz\">data.table_1.8.8.tar.gz</a> 2013-03-06 06:31 874K "
## [53] " <a href=\"data.table_1.9.2.tar.gz\">data.table_1.9.2.tar.gz</a> 2014-02-27 13:49 1.0M "
## [54] " <a href=\"data.table_1.9.4.tar.gz\">data.table_1.9.4.tar.gz</a> 2014-10-02 06:41 927K "
## [55] " <a href=\"data.table_1.9.6.tar.gz\">data.table_1.9.6.tar.gz</a> 2015-09-19 20:13 3.5M "
## [56] " <a href=\"data.table_1.9.8.tar.gz\">data.table_1.9.8.tar.gz</a> 2016-11-25 11:55 2.9M "
The output above shows 56 lines, which means there were some that were not captured by the regex. Which ones were they?
(missed.subjects <- grep(
nc::var_args_list(naive.pattern)$pattern,
tar.gz.vec, value=TRUE, invert=TRUE))
## [1] " <a href=\"data.table_1.10.4-1.tar.gz\">data.table_1.10.4-1.tar.gz</a> 2017-10-09 22:36 2.9M "
## [2] " <a href=\"data.table_1.10.4-2.tar.gz\">data.table_1.10.4-2.tar.gz</a> 2017-10-12 14:03 2.9M "
## [3] " <a href=\"data.table_1.10.4-3.tar.gz\">data.table_1.10.4-3.tar.gz</a> 2017-10-27 07:40 2.9M "
The output above indicates 3 missed subjects. Another way to determine we missed them would be to use that pattern as follows,
nc::capture_first_vec(tar.gz.vec, naive.pattern)
## Error in stop_for_na(make.na): subject(s) 5,6,7 (3 total) did not match regex below; to output missing rows use nomatch.error=FALSE
## (?:(?:_([0-9.]+)[.]tar[.]gz</a>\s+(.*?)\s))
tar.gz.vec[c(5,6,7)]
## [1] " <a href=\"data.table_1.10.4-1.tar.gz\">data.table_1.10.4-1.tar.gz</a> 2017-10-09 22:36 2.9M "
## [2] " <a href=\"data.table_1.10.4-2.tar.gz\">data.table_1.10.4-2.tar.gz</a> 2017-10-12 14:03 2.9M "
## [3] " <a href=\"data.table_1.10.4-3.tar.gz\">data.table_1.10.4-3.tar.gz</a> 2017-10-27 07:40 2.9M "
Below we revise the pattern so that it matches all of the tar files, including those with a hyphen in the version,
revised.pattern <- list(
"_",
version="[0-9.-]+",
"[.]tar[.]gz</a>\\s+",
date.str=".*?",
"\\s")
nc::capture_first_vec(tar.gz.vec, revised.pattern)
## version date.str
## <char> <char>
## 1: 1.0 2006-04-14
## 2: 1.1 2008-08-27
## 3: 1.10.0 2016-12-03
## 4: 1.10.2 2017-01-31
## 5: 1.10.4-1 2017-10-09
## 6: 1.10.4-2 2017-10-12
## 7: 1.10.4-3 2017-10-27
## 8: 1.10.4 2017-02-01
## 9: 1.11.0 2018-05-01
## 10: 1.11.2 2018-05-08
## 11: 1.11.4 2018-05-27
## 12: 1.11.6 2018-09-19
## 13: 1.11.8 2018-09-30
## 14: 1.12.0 2019-01-13
## 15: 1.12.2 2019-04-07
## 16: 1.12.4 2019-10-03
## 17: 1.12.6 2019-10-18
## 18: 1.12.8 2019-12-09
## 19: 1.13.0 2020-07-24
## 20: 1.13.2 2020-10-19
## 21: 1.13.4 2020-12-08
## 22: 1.13.6 2020-12-30
## 23: 1.14.0 2021-02-21
## 24: 1.2 2008-09-01
## 25: 1.4.1 2010-05-03
## 26: 1.5.1 2011-01-08
## 27: 1.5.2 2011-01-21
## 28: 1.5.3 2011-02-11
## 29: 1.5 2010-09-14
## 30: 1.6.1 2011-06-29
## 31: 1.6.2 2011-07-02
## 32: 1.6.3 2011-08-04
## 33: 1.6.4 2011-08-10
## 34: 1.6.5 2011-08-25
## 35: 1.6.6 2011-08-25
## 36: 1.6 2011-04-24
## 37: 1.7.1 2011-10-22
## 38: 1.7.10 2012-02-07
## 39: 1.7.2 2011-11-07
## 40: 1.7.3 2011-11-25
## 41: 1.7.4 2011-11-29
## 42: 1.7.5 2011-12-04
## 43: 1.7.6 2011-12-13
## 44: 1.7.7 2011-12-15
## 45: 1.7.8 2012-01-25
## 46: 1.7.9 2012-01-31
## 47: 1.8.0 2012-07-16
## 48: 1.8.10 2013-09-03
## 49: 1.8.2 2012-07-17
## 50: 1.8.4 2012-11-09
## 51: 1.8.6 2012-11-13
## 52: 1.8.8 2013-03-06
## 53: 1.9.2 2014-02-27
## 54: 1.9.4 2014-10-02
## 55: 1.9.6 2015-09-19
## 56: 1.9.8 2016-11-25
## version date.str
Analyze several packages for comparison
The code below defines a set of four packages for which we would like to analyze the release history (tidyverse packages for comparison).
library(data.table)
compare.pkg.dt <- rbind(
data.table(project="tidyverse", Package=c("readr","tidyr","dplyr")),
data.table(project="deprecated", Package=c("reshape2", "plyr")),
data.table(project="data.table", Package="data.table"))
In the code below, we do the same thing for each package,
(release.dt <- compare.pkg.dt[, {
Archive.pkg <- get_Archive(Package)
nc::capture_all_str(Archive.pkg, revised.pattern)
}, by=names(compare.pkg.dt)])
## project Package version date.str
## <char> <char> <char> <char>
## 1: tidyverse readr 0.1.0 2015-04-08
## 2: tidyverse readr 0.1.1 2015-05-29
## 3: tidyverse readr 0.2.0 2015-10-20
## 4: tidyverse readr 0.2.1 2015-10-21
## 5: tidyverse readr 0.2.2 2015-10-22
## ---
## 179: data.table data.table 1.8.8 2013-03-06
## 180: data.table data.table 1.9.2 2014-02-27
## 181: data.table data.table 1.9.4 2014-10-02
## 182: data.table data.table 1.9.6 2015-09-19
## 183: data.table data.table 1.9.8 2016-11-25
The result above is a data table with one row for each package
version. Note that the code set by to all column names, so that the
code is run for each row/package.
Add columns for plotting
For plotting we add a few more columns,
release.dt[, `:=`(
IDate = as.IDate(date.str),
year = as.integer(sub("-.*", "", date.str)),
package = factor(Package, compare.pkg.dt$Package),
Project = paste0('\n', project))]
setkey(release.dt, Project, Package, IDate)
release.dt
## project Package version date.str IDate year package Project
## <char> <char> <char> <char> <IDat> <int> <fctr> <char>
## 1: data.table data.table 1.0 2006-04-14 2006-04-14 2006 data.table \ndata.table
## 2: data.table data.table 1.1 2008-08-27 2008-08-27 2008 data.table \ndata.table
## 3: data.table data.table 1.2 2008-09-01 2008-09-01 2008 data.table \ndata.table
## 4: data.table data.table 1.4.1 2010-05-03 2010-05-03 2010 data.table \ndata.table
## 5: data.table data.table 1.5 2010-09-14 2010-09-14 2010 data.table \ndata.table
## ---
## 179: tidyverse tidyr 1.1.1 2020-07-31 2020-07-31 2020 tidyr \ntidyverse
## 180: tidyverse tidyr 1.1.2 2020-08-27 2020-08-27 2020 tidyr \ntidyverse
## 181: tidyverse tidyr 1.1.3 2021-03-03 2021-03-03 2021 tidyr \ntidyverse
## 182: tidyverse tidyr 1.1.4 2021-09-27 2021-09-27 2021 tidyr \ntidyverse
## 183: tidyverse tidyr 1.2.0 2022-02-01 2022-02-01 2022 tidyr \ntidyverse
To explain the new columns above,
IDateis for the date to display on the X axis,yearis for labeling the first released version each year,packageis for displaying the Y axis in a particular order (defined by the factor levels),Projectis for the facet/panel titles (newline so that minimal vertical space is used).
Basic plot
The code below creates a basic version history plot,
library(ggplot2)
## Warning: replacing previous import 'lifecycle::last_warnings' by 'rlang::last_warnings' when loading 'tibble'
## Warning: replacing previous import 'lifecycle::last_warnings' by 'rlang::last_warnings' when loading 'pillar'
(gg.points <- ggplot()+
theme(
axis.text.x=element_text(hjust=1, angle=40))+
facet_grid(Project ~ ., labeller=label_both, scales="free")+
geom_point(aes(
IDate, package),
shape=1,
data=release.dt)+
coord_cartesian(expand=5)+
scale_x_date("Date", breaks="year"))
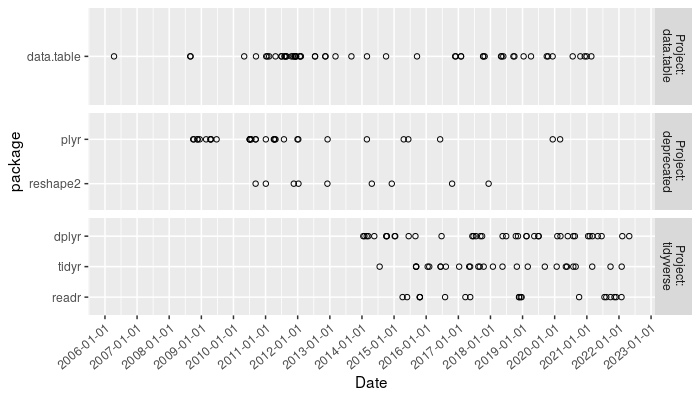
The plot above shows a point for every release to CRAN, so you can see the distribution of releases over time.
Add direct labels
Before plotting we make a new table which contains only the first
release of data.table in each year (for direct labels),
(labeled.releases <- release.dt[Package=="data.table", .SD[1], by=year])
## year project Package version date.str IDate package Project
## <int> <char> <char> <char> <char> <IDat> <fctr> <char>
## 1: 2006 data.table data.table 1.0 2006-04-14 2006-04-14 data.table \ndata.table
## 2: 2008 data.table data.table 1.1 2008-08-27 2008-08-27 data.table \ndata.table
## 3: 2010 data.table data.table 1.4.1 2010-05-03 2010-05-03 data.table \ndata.table
## 4: 2011 data.table data.table 1.5.1 2011-01-08 2011-01-08 data.table \ndata.table
## 5: 2012 data.table data.table 1.7.8 2012-01-25 2012-01-25 data.table \ndata.table
## 6: 2013 data.table data.table 1.8.8 2013-03-06 2013-03-06 data.table \ndata.table
## 7: 2014 data.table data.table 1.9.2 2014-02-27 2014-02-27 data.table \ndata.table
## 8: 2015 data.table data.table 1.9.6 2015-09-19 2015-09-19 data.table \ndata.table
## 9: 2016 data.table data.table 1.9.8 2016-11-25 2016-11-25 data.table \ndata.table
## 10: 2017 data.table data.table 1.10.2 2017-01-31 2017-01-31 data.table \ndata.table
## 11: 2018 data.table data.table 1.11.0 2018-05-01 2018-05-01 data.table \ndata.table
## 12: 2019 data.table data.table 1.12.0 2019-01-13 2019-01-13 data.table \ndata.table
## 13: 2020 data.table data.table 1.13.0 2020-07-24 2020-07-24 data.table \ndata.table
## 14: 2021 data.table data.table 1.14.0 2021-02-21 2021-02-21 data.table \ndata.table
gg.points+
directlabels::geom_dl(aes(
IDate, package, label=paste0(year, "\n", version)),
method=list(
cex=0.7,
directlabels::polygon.method(
"top", offset.cm=0.2, custom.colors=list(
colour="white",
box.color="black",
text.color="black"))),
data=labeled.releases)
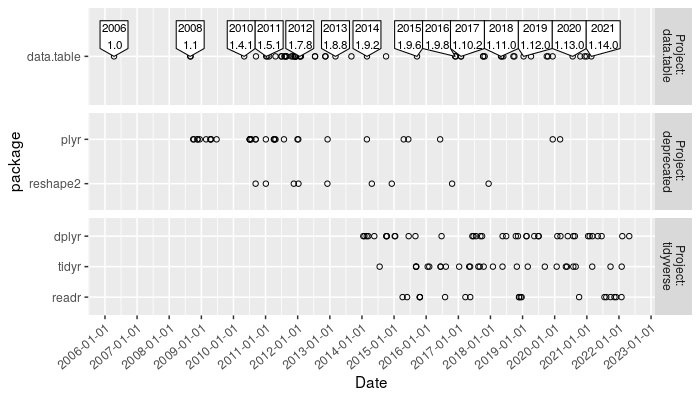
The plot above shows a label for the first version released each year.
Releases per year
One way to compute releases per year would be to add up the total number of releases, then divide by the number of years,
(overall.stats <- dcast(
release.dt,
project + Package ~ .,
list(min,max,length),
value.var="year"
)[, releases.per.year := year_length/(year_max-year_min+1)][])
## Warning in dcast.data.table(release.dt, project + Package ~ ., list(min, : NAs introduced by coercion to integer range
## Warning in dcast.data.table(release.dt, project + Package ~ ., list(min, : NAs introduced by coercion to integer range
## project Package year_min year_max year_length releases.per.year
## <char> <char> <int> <int> <int> <num>
## 1: data.table data.table 2006 2021 56 3.500000
## 2: deprecated plyr 2008 2020 32 2.461538
## 3: deprecated reshape2 2010 2017 9 1.125000
## 4: tidyverse dplyr 2014 2022 39 4.333333
## 5: tidyverse readr 2015 2022 19 2.375000
## 6: tidyverse tidyr 2014 2022 28 3.111111
Another way to do it would be to compute the number of releases in each year since the release of the package. To do that we first compute, for each package, a set of years for which we want to count releases.
(max.year <- max(release.dt$year))
## [1] 2022
(years.since.release <- release.dt[, .(
year=seq(min(year), max.year)
), by=.(Project, project, Package, package)])
## Project project Package package year
## <char> <char> <char> <fctr> <int>
## 1: \ndata.table data.table data.table data.table 2006
## 2: \ndata.table data.table data.table data.table 2007
## 3: \ndata.table data.table data.table data.table 2008
## 4: \ndata.table data.table data.table data.table 2009
## 5: \ndata.table data.table data.table data.table 2010
## 6: \ndata.table data.table data.table data.table 2011
## 7: \ndata.table data.table data.table data.table 2012
## 8: \ndata.table data.table data.table data.table 2013
## 9: \ndata.table data.table data.table data.table 2014
## 10: \ndata.table data.table data.table data.table 2015
## 11: \ndata.table data.table data.table data.table 2016
## 12: \ndata.table data.table data.table data.table 2017
## 13: \ndata.table data.table data.table data.table 2018
## 14: \ndata.table data.table data.table data.table 2019
## 15: \ndata.table data.table data.table data.table 2020
## 16: \ndata.table data.table data.table data.table 2021
## 17: \ndata.table data.table data.table data.table 2022
## 18: \ndeprecated deprecated plyr plyr 2008
## 19: \ndeprecated deprecated plyr plyr 2009
## 20: \ndeprecated deprecated plyr plyr 2010
## 21: \ndeprecated deprecated plyr plyr 2011
## 22: \ndeprecated deprecated plyr plyr 2012
## 23: \ndeprecated deprecated plyr plyr 2013
## 24: \ndeprecated deprecated plyr plyr 2014
## 25: \ndeprecated deprecated plyr plyr 2015
## 26: \ndeprecated deprecated plyr plyr 2016
## 27: \ndeprecated deprecated plyr plyr 2017
## 28: \ndeprecated deprecated plyr plyr 2018
## 29: \ndeprecated deprecated plyr plyr 2019
## 30: \ndeprecated deprecated plyr plyr 2020
## 31: \ndeprecated deprecated plyr plyr 2021
## 32: \ndeprecated deprecated plyr plyr 2022
## 33: \ndeprecated deprecated reshape2 reshape2 2010
## 34: \ndeprecated deprecated reshape2 reshape2 2011
## 35: \ndeprecated deprecated reshape2 reshape2 2012
## 36: \ndeprecated deprecated reshape2 reshape2 2013
## 37: \ndeprecated deprecated reshape2 reshape2 2014
## 38: \ndeprecated deprecated reshape2 reshape2 2015
## 39: \ndeprecated deprecated reshape2 reshape2 2016
## 40: \ndeprecated deprecated reshape2 reshape2 2017
## 41: \ndeprecated deprecated reshape2 reshape2 2018
## 42: \ndeprecated deprecated reshape2 reshape2 2019
## 43: \ndeprecated deprecated reshape2 reshape2 2020
## 44: \ndeprecated deprecated reshape2 reshape2 2021
## 45: \ndeprecated deprecated reshape2 reshape2 2022
## 46: \ntidyverse tidyverse dplyr dplyr 2014
## 47: \ntidyverse tidyverse dplyr dplyr 2015
## 48: \ntidyverse tidyverse dplyr dplyr 2016
## 49: \ntidyverse tidyverse dplyr dplyr 2017
## 50: \ntidyverse tidyverse dplyr dplyr 2018
## 51: \ntidyverse tidyverse dplyr dplyr 2019
## 52: \ntidyverse tidyverse dplyr dplyr 2020
## 53: \ntidyverse tidyverse dplyr dplyr 2021
## 54: \ntidyverse tidyverse dplyr dplyr 2022
## 55: \ntidyverse tidyverse readr readr 2015
## 56: \ntidyverse tidyverse readr readr 2016
## 57: \ntidyverse tidyverse readr readr 2017
## 58: \ntidyverse tidyverse readr readr 2018
## 59: \ntidyverse tidyverse readr readr 2019
## 60: \ntidyverse tidyverse readr readr 2020
## 61: \ntidyverse tidyverse readr readr 2021
## 62: \ntidyverse tidyverse readr readr 2022
## 63: \ntidyverse tidyverse tidyr tidyr 2014
## 64: \ntidyverse tidyverse tidyr tidyr 2015
## 65: \ntidyverse tidyverse tidyr tidyr 2016
## 66: \ntidyverse tidyverse tidyr tidyr 2017
## 67: \ntidyverse tidyverse tidyr tidyr 2018
## 68: \ntidyverse tidyverse tidyr tidyr 2019
## 69: \ntidyverse tidyverse tidyr tidyr 2020
## 70: \ntidyverse tidyverse tidyr tidyr 2021
## 71: \ntidyverse tidyverse tidyr tidyr 2022
## Project project Package package year
Then we can do a join and summarize to count the number of releases in each year, for each package,
(releases.per.year <- release.dt[years.since.release, .(
N=as.numeric(.N)
), on=.NATURAL, by=.EACHI])
## project Package year package Project N
## <char> <char> <int> <fctr> <char> <num>
## 1: data.table data.table 2006 data.table \ndata.table 1
## 2: data.table data.table 2007 data.table \ndata.table 0
## 3: data.table data.table 2008 data.table \ndata.table 2
## 4: data.table data.table 2009 data.table \ndata.table 0
## 5: data.table data.table 2010 data.table \ndata.table 2
## 6: data.table data.table 2011 data.table \ndata.table 17
## 7: data.table data.table 2012 data.table \ndata.table 7
## 8: data.table data.table 2013 data.table \ndata.table 2
## 9: data.table data.table 2014 data.table \ndata.table 2
## 10: data.table data.table 2015 data.table \ndata.table 1
## 11: data.table data.table 2016 data.table \ndata.table 2
## 12: data.table data.table 2017 data.table \ndata.table 5
## 13: data.table data.table 2018 data.table \ndata.table 5
## 14: data.table data.table 2019 data.table \ndata.table 5
## 15: data.table data.table 2020 data.table \ndata.table 4
## 16: data.table data.table 2021 data.table \ndata.table 1
## 17: data.table data.table 2022 data.table \ndata.table 0
## 18: deprecated plyr 2008 plyr \ndeprecated 5
## 19: deprecated plyr 2009 plyr \ndeprecated 5
## 20: deprecated plyr 2010 plyr \ndeprecated 7
## 21: deprecated plyr 2011 plyr \ndeprecated 7
## 22: deprecated plyr 2012 plyr \ndeprecated 2
## 23: deprecated plyr 2013 plyr \ndeprecated 0
## 24: deprecated plyr 2014 plyr \ndeprecated 1
## 25: deprecated plyr 2015 plyr \ndeprecated 2
## 26: deprecated plyr 2016 plyr \ndeprecated 1
## 27: deprecated plyr 2017 plyr \ndeprecated 0
## 28: deprecated plyr 2018 plyr \ndeprecated 0
## 29: deprecated plyr 2019 plyr \ndeprecated 1
## 30: deprecated plyr 2020 plyr \ndeprecated 1
## 31: deprecated plyr 2021 plyr \ndeprecated 0
## 32: deprecated plyr 2022 plyr \ndeprecated 0
## 33: deprecated reshape2 2010 reshape2 \ndeprecated 1
## 34: deprecated reshape2 2011 reshape2 \ndeprecated 2
## 35: deprecated reshape2 2012 reshape2 \ndeprecated 2
## 36: deprecated reshape2 2013 reshape2 \ndeprecated 0
## 37: deprecated reshape2 2014 reshape2 \ndeprecated 2
## 38: deprecated reshape2 2015 reshape2 \ndeprecated 0
## 39: deprecated reshape2 2016 reshape2 \ndeprecated 1
## 40: deprecated reshape2 2017 reshape2 \ndeprecated 1
## 41: deprecated reshape2 2018 reshape2 \ndeprecated 0
## 42: deprecated reshape2 2019 reshape2 \ndeprecated 0
## 43: deprecated reshape2 2020 reshape2 \ndeprecated 0
## 44: deprecated reshape2 2021 reshape2 \ndeprecated 0
## 45: deprecated reshape2 2022 reshape2 \ndeprecated 0
## 46: tidyverse dplyr 2014 dplyr \ntidyverse 8
## 47: tidyverse dplyr 2015 dplyr \ntidyverse 4
## 48: tidyverse dplyr 2016 dplyr \ntidyverse 1
## 49: tidyverse dplyr 2017 dplyr \ntidyverse 5
## 50: tidyverse dplyr 2018 dplyr \ntidyverse 4
## 51: tidyverse dplyr 2019 dplyr \ntidyverse 5
## 52: tidyverse dplyr 2020 dplyr \ntidyverse 5
## 53: tidyverse dplyr 2021 dplyr \ntidyverse 5
## 54: tidyverse dplyr 2022 dplyr \ntidyverse 2
## 55: tidyverse readr 2015 readr \ntidyverse 5
## 56: tidyverse readr 2016 readr \ntidyverse 1
## 57: tidyverse readr 2017 readr \ntidyverse 2
## 58: tidyverse readr 2018 readr \ntidyverse 4
## 59: tidyverse readr 2019 readr \ntidyverse 0
## 60: tidyverse readr 2020 readr \ntidyverse 1
## 61: tidyverse readr 2021 readr \ntidyverse 5
## 62: tidyverse readr 2022 readr \ntidyverse 1
## 63: tidyverse tidyr 2014 tidyr \ntidyverse 1
## 64: tidyverse tidyr 2015 tidyr \ntidyverse 3
## 65: tidyverse tidyr 2016 tidyr \ntidyverse 5
## 66: tidyverse tidyr 2017 tidyr \ntidyverse 6
## 67: tidyverse tidyr 2018 tidyr \ntidyverse 3
## 68: tidyverse tidyr 2019 tidyr \ntidyverse 2
## 69: tidyverse tidyr 2020 tidyr \ntidyverse 5
## 70: tidyverse tidyr 2021 tidyr \ntidyverse 2
## 71: tidyverse tidyr 2022 tidyr \ntidyverse 1
## project Package year package Project N
Note that on=.NATURAL above means to join on the common columns
between the two tables, and by=.EACHI means to compute a summary for
each value specified in i (the first argument in the square bracket).
We can plot these data as a heat map via
ggplot()+
theme_bw()+
theme(panel.spacing=grid::unit(0, "lines"))+
geom_tile(aes(
year, package, fill=log(N+1)),
data=releases.per.year)+
geom_text(aes(
year, package, label=N),
data=releases.per.year)+
facet_grid(Project ~ ., labeller=label_both, scales="free", space="free")+
scale_fill_gradient("releases\n(log scale)", low="white", high="red")+
scale_x_continuous(breaks=seq(2006, 2022, by=2))+
coord_cartesian(expand=FALSE)
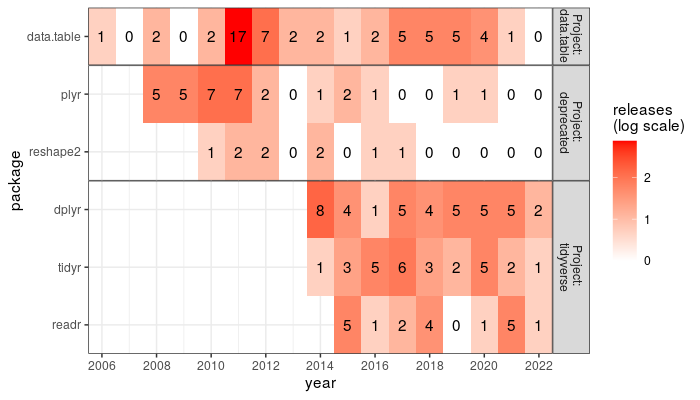
The heat map above shows a summarized display of the release data we saw earlier in the dot plot.
Next, we can apply a list of summary functions over all of the yearly counts, for each package, via
(per.year.stats <- dcast(
releases.per.year,
project + Package ~ .,
list(min, max, mean, sd, length),
value.var = "N"))
## project Package N_min N_max N_mean N_sd N_length
## <char> <char> <num> <num> <num> <num> <int>
## 1: data.table data.table 0 17 3.2941176 4.0890816 17
## 2: deprecated plyr 0 7 2.1333333 2.5597619 15
## 3: deprecated reshape2 0 2 0.6923077 0.8548504 13
## 4: tidyverse dplyr 1 8 4.3333333 2.0000000 9
## 5: tidyverse readr 0 5 2.3750000 1.9955307 8
## 6: tidyverse tidyr 1 6 3.1111111 1.8333333 9
Finally, the code below creates a table to compare the two different ways of computing the number of releases per year,
per.year.stats[overall.stats, .(
Package,
overall.mean=N_mean,
mean.per.year=releases.per.year
), on="Package"]
## Package overall.mean mean.per.year
## <char> <num> <num>
## 1: data.table 3.2941176 3.500000
## 2: plyr 2.1333333 2.461538
## 3: reshape2 0.6923077 1.125000
## 4: dplyr 4.3333333 4.333333
## 5: readr 2.3750000 2.375000
## 6: tidyr 3.1111111 3.111111
The table above show similar numbers for the two methods of computing the number of releases per year.
Conclusion
We have shown how to download CRAN package release data, how to parse
the web pages using the nc package and a regular expression, how to
summarize/analyze using data.table, and how to visualize using
ggplot2.