Seasonal temperature variations where I have lived
I used this R script to make the plot below, which shows averages of daily high and low temperatures, for each month, in each city where I have lived.
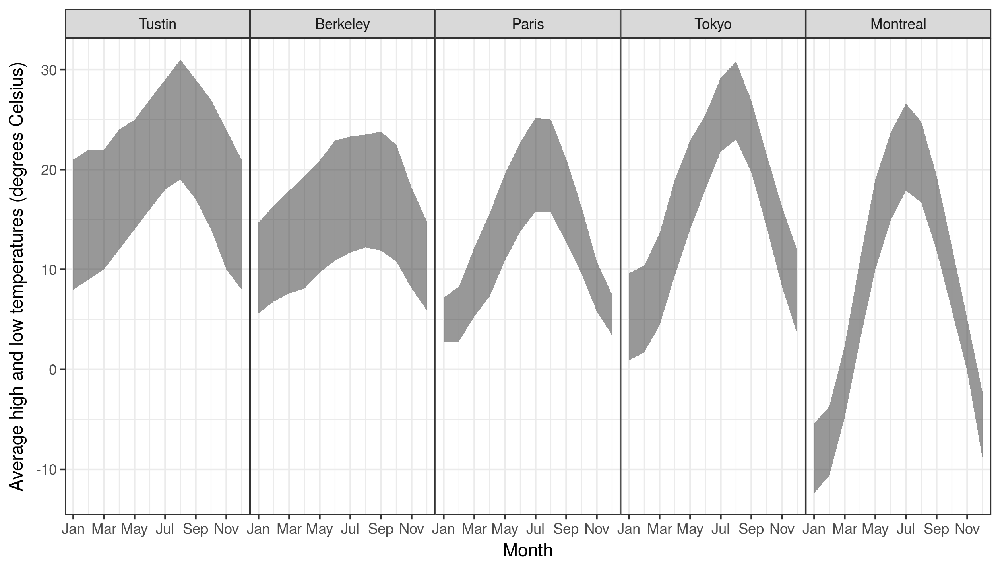
I got the data from wikipedia pages, defined in a named character vector in R (the names are used for the panel titles in the plot):
url.vec <- c(
Tustin="https://en.wikipedia.org/wiki/Tustin,_California",
Berkeley="https://en.wikipedia.org/wiki/Berkeley,_California",
Paris="https://en.wikipedia.org/wiki/Paris",
Tokyo="https://en.wikipedia.org/wiki/Tokyo",
Montreal="https://en.wikipedia.org/wiki/Montreal")
The nice thing about my script is that to add a new city, all I have to do is add one named element to the character vector above. And then the code below takes care of all the downloading, processing, and plotting. It works for the few cities where I have lived, but I wonder if it would work on all wikipedia city pages? I’ll leave that for future work.
For each city, I used the excellent download.file function to get
a local copy of the wikipedia web page:
city.html <- file.path("wikipedia", paste0(city, ".html"))
if(!file.exists(city.html)){
u <- url.vec[[city]]
download.file(u, city.html)
}
Then I used htmltab to extract the table of temperature data from the web page (note the XPath expression which means to look for the table element with a Month th element):
df <- htmltab(
city.html,
which="//th[text()='Month']/ancestor::table")
That gives us a data.frame with character columns:
> str(df)
'data.frame': 13 obs. of 14 variables:
$ Climate data for McGill University (McTavish), 1971–2000 normals, extremes 1871–present >> Month: chr "Record high °C (°F)" "Average high °C (°F)" "Daily mean °C (°F)" "Average low °C (°F)" ...
$ Climate data for McGill University (McTavish), 1971–2000 normals, extremes 1871–present >> Jan : chr "12.8 (55)" "−5.4 (22.3)" "−8.9 (16)" "−12.4 (9.7)" ...
$ Climate data for McGill University (McTavish), 1971–2000 normals, extremes 1871–present >> Feb : chr "15.0 (59)" "−3.7 (25.3)" "−7.2 (19)" "−10.6 (12.9)" ...
>
We can get the months by deleting all the text in the column names except the last word:
> col.name.vec <- sub(".*> ", "", names(df))
> col.name.vec
[1] "Month" "Jan" "Feb" "Mar" "Apr" "May" "Jun" "Jul" "Aug"
[10] "Sep" "Oct" "Nov" "Dec" "Year"
>
The first column of the data.frame contains the climate variable names:
> row.indices <- -grep("^Source", df[,1])
> row.name.vec <- df[row.indices, 1]
> row.name.vec
[1] "Record high °C (°F)"
[2] "Average high °C (°F)"
[3] "Daily mean °C (°F)"
[4] "Average low °C (°F)"
[5] "Record low °C (°F)"
[6] "Average precipitation mm (inches)"
[7] "Average rainfall mm (inches)"
[8] "Average snowfall cm (inches)"
[9] "Average precipitation days (≥ 0.2 mm)"
[10] "Average rainy days (≥ 0.2 mm)"
[11] "Average snowy days (≥ 0.2 cm)"
[12] "Mean monthly sunshine hours"
>
But note that different web pages have different kinds of climate data tables. The table above is for Montreal – in Canada the metric system is used, so Celsius degrees are listed first. However in the USA we have fewer variables, and Fahrenheit is listed first:
> city <- "Berkeley"
> city.html <- file.path("wikipedia", paste0(city, ".html"))
> if(!file.exists(city.html)){
+ u <- url.vec[[city]]
+ download.file(u, city.html)
+ }
> df <- htmltab(city.html, which="//th[text()='Month']/ancestor::table")
> col.name.vec <- sub(".*> ", "", names(df))
> row.indices <- -grep("^Source", df[,1])
> row.name.vec <- df[row.indices, 1]
> row.name.vec
[1] "Record high °F (°C)" "Average high °F (°C)"
[3] "Average low °F (°C)" "Record low °F (°C)"
[5] "Average precipitation inches (mm)"
>
The following regular expression can be used to extract the variable name, the unit before the parentheses, and the unit inside the parentheses.
pattern <- paste0(
"(?<varname>.*)",
" ",
"(?<before>[^ (]+)",
" [(]",
"(?<inside>[^)]+)",
"[)]")
We can extract the data using the namedCapture::str_match_named function:
> row.name.mat <- str_match_named(row.name.vec, pattern)
> row.name.mat
varname before inside
[1,] "Record high" "°F" "°C"
[2,] "Average high" "°F" "°C"
[3,] "Average low" "°F" "°C"
[4,] "Record low" "°F" "°C"
[5,] "Average precipitation" "inches" "mm"
>
We have an if statement that checks for °C inside or before the parentheses, and uses the correpsonding column:
is.C <- row.name.mat=="°C"
keep.name <- if(any(is.C[, "inside"], na.rm=TRUE)){
"inside"
}else if(any(is.C[, "before"], na.rm=TRUE)){
"before"
}else{
stop("no °C in or before parentheses")
}
We also use the regex to extract the data:
> chr.mat <- unname(as.matrix(df[row.indices, col.indices]))
> data.match.mat <- str_match_named(paste0(" ", chr.mat), pattern, list(
+ before=to.numeric,
+ inside=to.numeric))
> head(data.match.mat)
varname before inside
1 77.00 25.0
2 58.40 14.7
3 42.00 5.6
4 25.00 -4.0
5 4.98 126.5
6 80.00 27.0
>
Note the third argument above, which is a list of conversion functions (which take as input one of the matched groups as a character vector). Here we use the custom to.numeric function because there are some commas and non-standard dashes which we must remove in order for the standard as.numeric function to work:
to.numeric <- function(chr.vec){
as.numeric(gsub(",", "", gsub("−", "-", chr.vec)))
}
After a few more conversions (see code for details), we get the following wide data.table:
> wide.dt <- data.table(
+ month=factor(month.vec, month.vec),
+ t(matrix(
+ to.numeric(chr.mat), nrow(chr.mat), ncol(chr.mat),
+ dimnames=list(new.row.names))))
> wide.dt
month Record high Average high Average low Record low Average precipitation
1: Jan 25 14.7 5.6 -4 126.5
2: Feb 27 16.4 6.8 -2 132.3
3: Mar 31 17.9 7.6 1 98.0
4: Apr 35 19.3 8.1 2 42.2
5: May 38 20.9 9.7 2 21.8
6: Jun 42 22.9 10.9 4 3.8
7: Jul 42 23.3 11.7 4 0.3
8: Aug 40 23.5 12.2 6 1.5
9: Sep 41 23.8 11.9 3 6.1
10: Oct 37 22.5 10.8 3 34.8
11: Nov 30 18.2 8.1 1 83.8
12: Dec 27 14.8 5.9 -4 128.0
>
We combine the data from the different cities using the rbind-list-of-data-tables idiom, we consider only the Average high and low variables, then we create a wide data.table using dcast:
> climate.dt <- data.table(do.call(rbind, climate.dt.list))
> temp.dt <- climate.dt[variable %in% c("Average high", "Average low")]
> temp.wide <- dcast(temp.dt, city + month ~ variable)
> head(temp.wide)
city month Average high Average low
1: Tustin Jan 21 8
2: Tustin Feb 22 9
3: Tustin Mar 22 10
4: Tustin Apr 24 12
5: Tustin May 25 14
6: Tustin Jun 27 16
>
This data.table can be plotted using geom_ribbon:
ggplot()+
geom_ribbon(aes(as.numeric(month), ymax=`Average high`, ymin=`Average low`),
alpha=0.5,
data=temp.wide)+
theme_bw()+
theme(panel.margin=grid::unit(0, "lines"))+
facet_grid(. ~ city)+
scale_x_continuous(
"Month",
breaks=breaks.vec,
labels=levels(temp.wide$month)[breaks.vec])+
scale_y_continuous(
"Average high and low temperatures (degrees Celsius)")